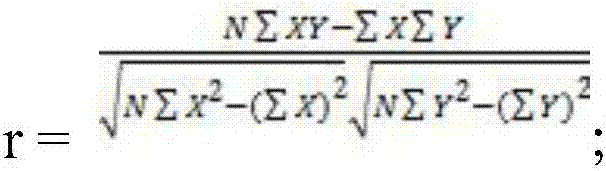Prediction method for target gene of long-fragment non-coding RNA of forest tree
A prediction method and target gene technology, applied in the field of molecular genetics, can solve the problems of false positives of target genes, low threshold parameters of prediction methods, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] The target gene of populus tomentosa lncRNA lnc-CK is predicted by using the method for predicting the target gene of forest long fragment non-coding RNA of the present invention.
[0049] 1. Obtaining lnc-CK sequence. The leaves of the Populus tomentosa clone "LM50" were collected, and the total RNA of the leaves of Populus tomentosa was extracted by using the CTAB method. After the quality assessment, it was sent to the company for sequencing analysis, and the sequence data of lncRNA expressed in the leaves of Populus tomentosa were obtained. Then the sequence of Populus tomentosa lncRNA lnc-CK, SEQ ID NO.1, was obtained.
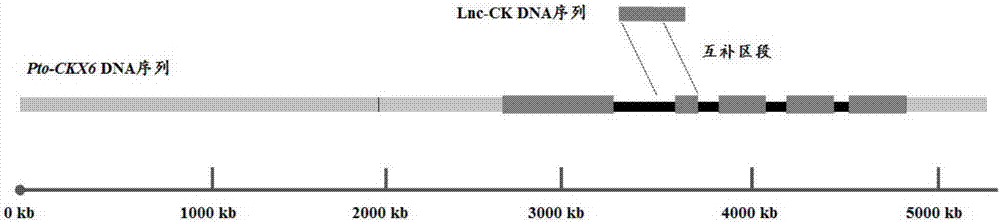
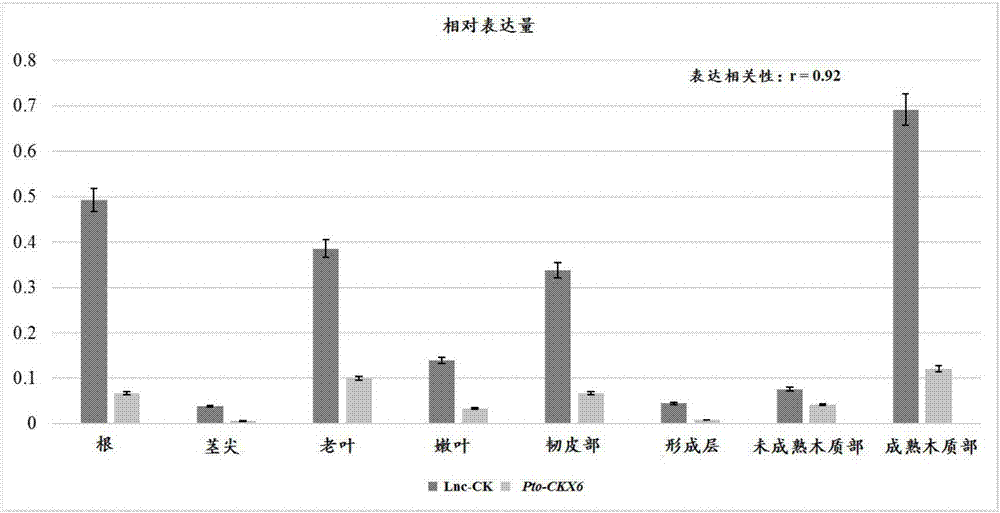
[0050]2. Preliminary prediction of the target gene of the lncRNA obtained in step 1 by using the prediction method of blast and RNAplex in combination. First, use the method of blast to compare lnc-CK with the sequences from the cDNA library of Populus tomentosa leaf tissue (E-valuefigure 1 shown. Preliminary screening obtained Pto-CKX6 as its pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com