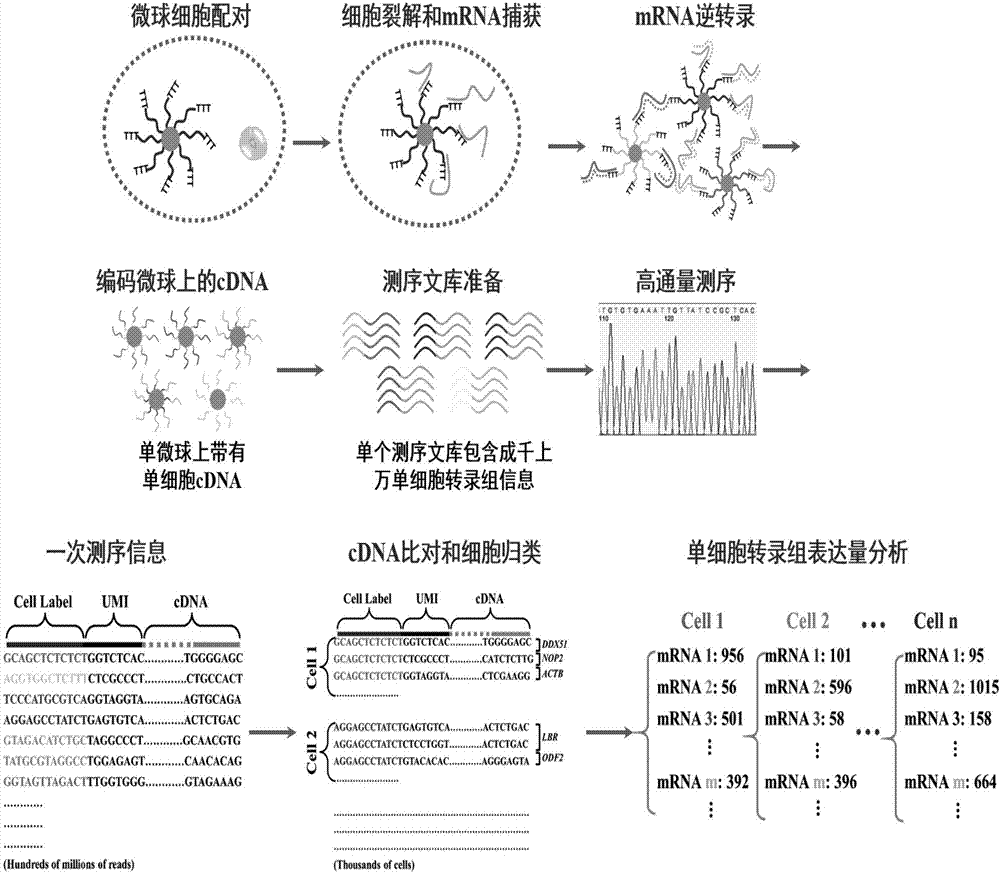
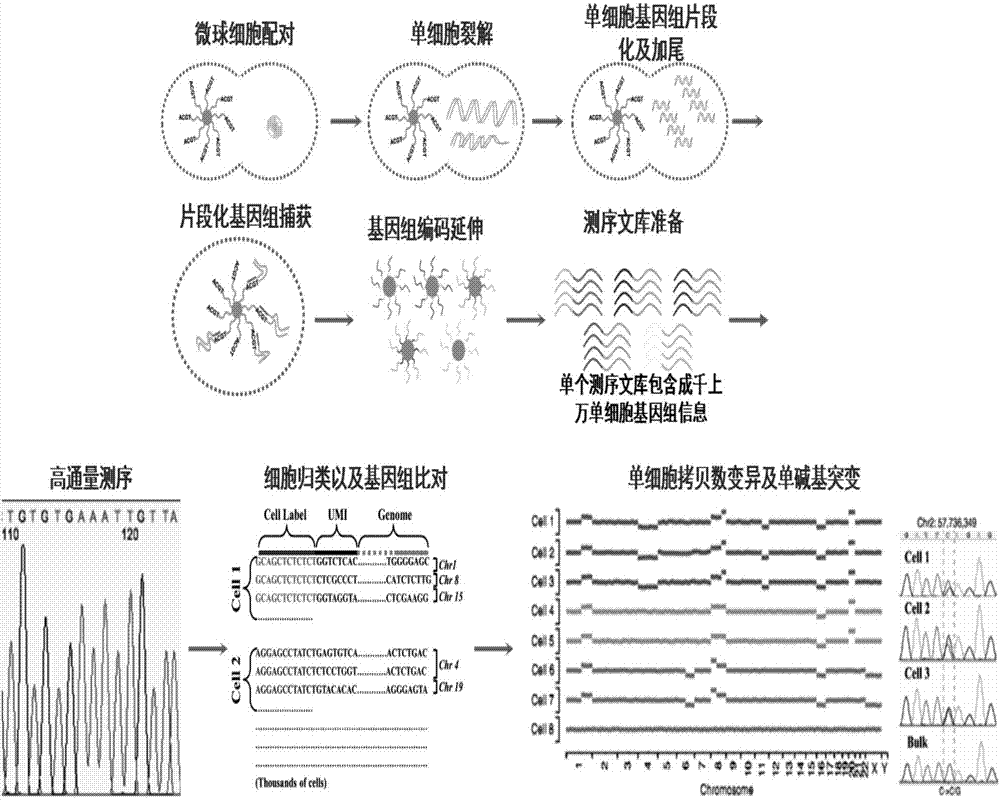
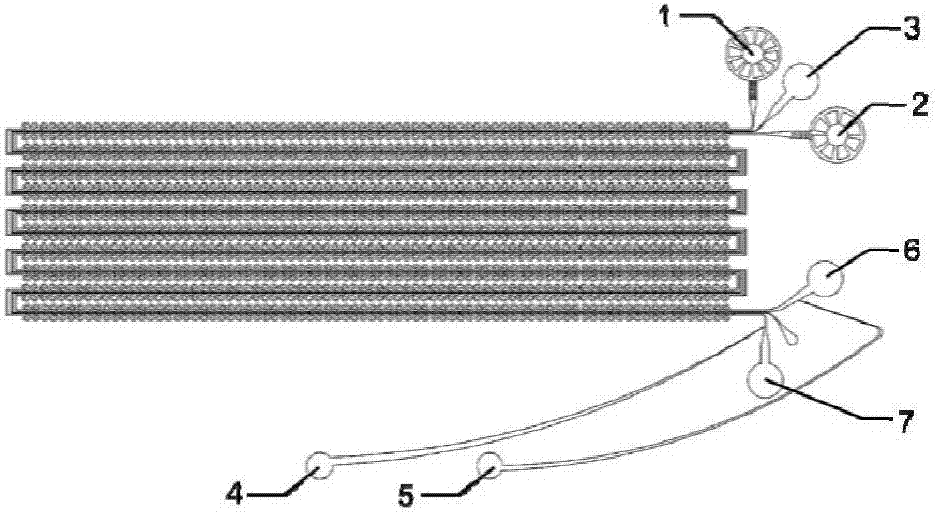
Method for high-throughput analysis of single cell inclusions by using paired micro-fluidic chip
A technology of microfluidic chips and inclusions, applied in the field of molecular and cell biology, can solve the problems that high-throughput analysis is not involved, single-cell inclusions cannot be analyzed, effective analysis targets are reduced, and the occupancy rate is achieved. High, high pairing efficiency, high-throughput effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Such as Figure 1 to Figure 3 As shown, the present invention is used for high-throughput paired capture of single microspheres and single cells. The chip is processed by standard soft lithography technology and includes three parts: a capture layer, a control layer and a slide. The capture layer is composed of three parts: cell flow channel (10), microsphere flow channel (11) and connection channel (13), and the control layer is composed of isolation channel (12). Wherein the cell flow path contains a plurality of cell capture units (8) connected in series, and each unit consists of three parts: a flow path (10), a liquid storage chamber (14), and a capture channel (16); see image 3 , the flow path (10) comprises a U-shaped tube, the left arm end of the U-shaped tube of the previous unit is connected to the right arm end of the U-shaped tube of the next unit, the connecting part between the U-shaped tubes is a straight tube, and the liquid storage chamber The chamber...
Embodiment 2
[0062] High-throughput single-cell transcriptome analysis
[0063]a. Single cell capture: Fill the isolation pump inlet (3) with solution, increase the pressure of the syringe pump, the isolation pump (12) deforms, and the diaphragm (18) squeezes the uppermost layer of the connecting channel until the connecting channel (13) is completely blocked Open, pass through the cell A549 suspension from the channel inlet (1) with a syringe pump, when the single cell enters the capture unit (8), because the path that the fluid passes through from the flow channel (10) is greater than from the liquid storage chamber (14), The cells will enter the liquid storage chamber (14) first, because the capture channel (16) is smaller than the cells, the cells are stuck in front of the capture channel (16), and at the same time block the capture channel (16), and flow through the liquid storage chamber (14) The fluid flow rate approaches zero, and at this time, subsequent cells cannot enter the liq...
Embodiment 3
[0087] Example 3: Single-cell genome analysis
[0088] High-throughput single-cell genome analysis
[0089] a. Single cell capture: Fill the isolation pump inlet (3) with solution, increase the pressure of the syringe pump, the isolation pump (12) deforms, and the diaphragm (18) squeezes the uppermost layer of the connecting channel until the connecting channel (13) is completely blocked Open, pass through the cell A549 suspension from the channel inlet (1) with a syringe pump, when the single cell enters the capture unit (8), because the path that the fluid passes through from the flow channel (10) is greater than from the liquid storage chamber (14), The cells will enter the liquid storage chamber (14) first, because the capture channel (16) is smaller than the cells, the cells are stuck in front of the capture channel (16), and at the same time block the capture channel (16), and flow through the liquid storage chamber (14) The fluid flow rate approaches zero, and at this ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com