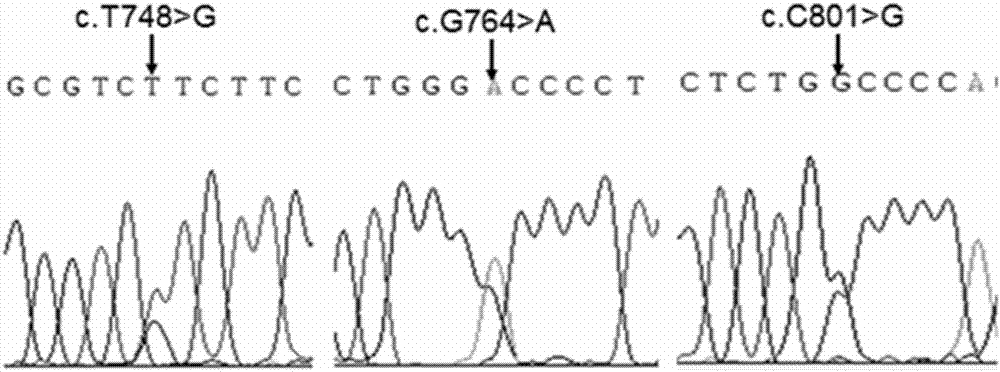
Three goat MC1R deficient mutants and application thereof
A technology for mutants and defects, applied in the field of goat molecular marker preparation, can solve problems such as impact
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Construction of eukaryotic expression vectors for wild-type goat melanocortin receptor-1 (gMC1R) and natural mutants
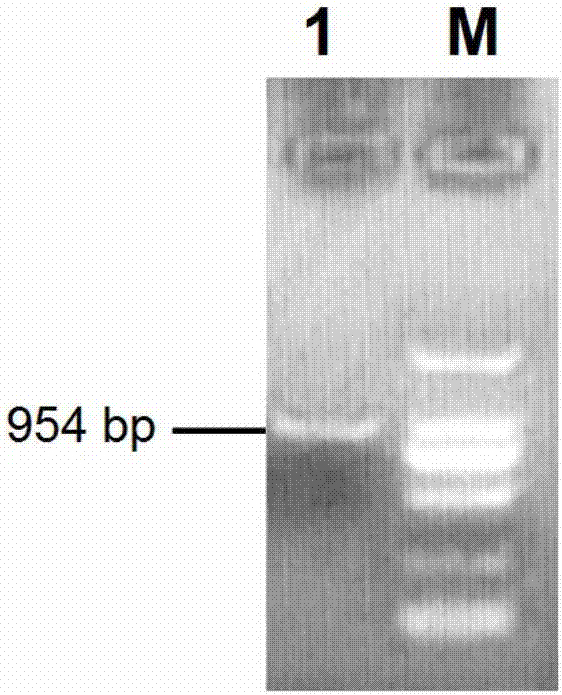
[0075] The results of bioinformatics analysis on gMC1R show that gMC1R has no intron, and the open reading frame of the gMC1R gene (GenBank accession number: XM_013970999.1) is 954bp, encoding 316 amino acids (see the sequence table SEQ ID NO: 1); The receptor protein has 7 transmembrane domains, 3 outer membrane loops and 3 membrane inner loops.
[0076] In this example, the eukaryotic expression vector pcDNA3.1 (purchased from Clontech) was used to construct the gMC1R wild-type plasmid and 8 mutant plasmids. The specific operation was as follows: using an upstream primer containing a Hind III restriction site and a Kozak sequence (CCCAAGCTTGCCACCATGCCTGCACTCGGCTCCC) and the downstream primer (CGCGGATCCTCACCAGGAGCACTGCAGCACC) containing the BamH I restriction site were used to amplify the coding region of the wild-type gMC1R gene by PCR. After loading ...
Embodiment 2
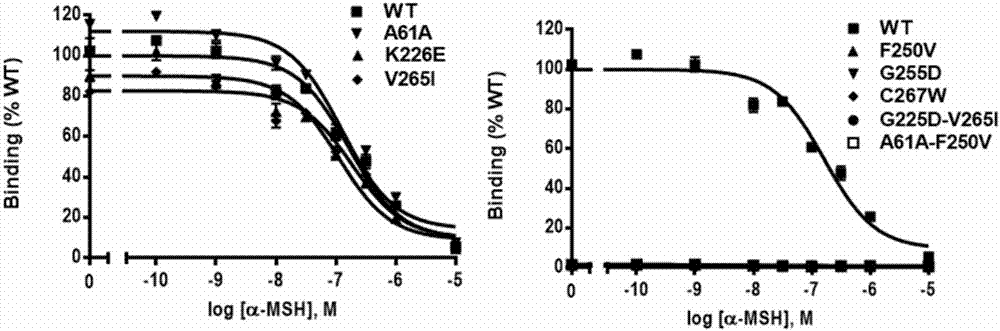
[0078] Ligand affinity test of wild-type gMC1R and mutants
[0079] Before the cells were collected, the cells were washed twice with serum-free medium (purchased from Gibco), and different final concentrations (10 -10 —10 -5 M) Ligand α-MSH (purchased from Bachem Company) and 100,000 cpm radiolabeled ligand 125 I-NDP-MSH (purchased from Peptide Radioactive Iodine Labeling Service Center of the University of Mississippi) (50 μL) was co-incubated at 37°C for 1 h at 37°C to transfect HEK293T cells (purchased from ATCC Company) with gMC1R wild receptors, and after the reaction was terminated, With pre-cooled PBS (137mM NaCl, 2.7mM KCl, 1.4mM KH 2 PO 4 , 4.3 mM Na 2 HPO 4 , pH 7.4) to wash off the unbound markers, add 100 μL of 0.5N NaOH to collect the cells, and use a gamma scintillation counter to measure the receptor binding in the cells 125 The total amount of I-NDP-MSH, that is, the binding ability of the MC1R mutant to the ligand α-MSH. GraphPad Prism5 software was us...
Embodiment 3
[0086] Comparison of cAMP activity of constitutive and inducible signaling molecules produced by wild-type gMC1R and mutants
[0087] Before collecting the cells, wash the cells twice with serum-free medium, and incubate the transfected gMC1R wild receptor and HEK293T cells of mutants were treated for 15min, and then different final concentrations (10 -10 —10 -5 M) α-MSH stimulated the treated cells, incubated at 37°C for 1 hour, put the cell culture plate on ice, discarded the medium, added 0.5N perchloric acid containing 180 μg / mL theophylline to extract intracellular cAMP, and irradiated Immunoassay (RIA) method 15 .Determination of the dose-dependent curve produced by the signal molecule cAMP, and using GraphPadPrism5 software to calculate its zero concentration value (Basal), effective intermediate concentration (EC 50 ) and the maximum concentration value (E max ).
[0088] The results showed that the mutants G255D, C267W and G225D-V265I almost completely lost the a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com