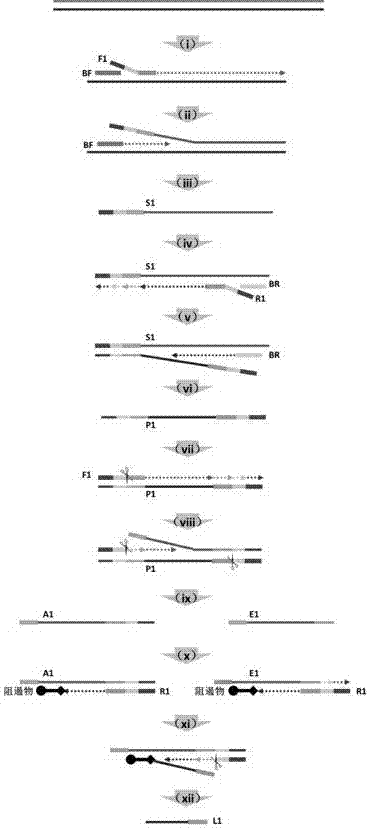
Method for generating single-stranded product in isothermal amplification system
A constant temperature amplification and product technology, applied in biochemical equipment and methods, microbial measurement/testing, DNA preparation, etc., can solve the problem of inconvenient capture of probes, achieve low cost, fast reaction rate, and overcome technical defects Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Steps for generating a 60nt single-stranded product at 56°C using the CaMV 35S of the transgenic rice KMD1 as a template.
[0035] CaMV 35S template sequence:
[0036] CACGTCTTCAAAGCAAGTGGATTGATGTGATATCTCCCACTGACGTAAGGGATGACGCACAATCCCACTATTCCTTCGCAAGACCTCTTCCTTATAAGGAAGTTCATTTCATTTGGAGAGGACACGCTGAAATCACCAGTCTCTCTCTAAATCTATCTCTCTC.
[0037] Complementary sequence of CaMV 35S template:
[0038] GAGAGAGATAGATTTGTAGAGAGAGACTGGTGATTTCACACATCAATCCACTTGCTTTGAAGACGTG.
[0039] Preparation of reagents before reaction: Add 50ul reaction reagents to the PCR tube: 10U Nt.BstNBI, 6U Bst DNAPolymerase, 2.5ul NEBuffer 3, 5ul Thermopol Reaction Buffer, 300uM dNTP, 500nM primers F1 and R1, 50nM primers BF and BR, 2.5 ul template, 50 nM nucleic acid repressor. Incubate at 56°C for 10 minutes.
[0040] The sequences of primers and nucleic acid repressors designed for the CaMV 35S double-stranded template sequence are as follows:
[0041] F1: CTTATTGTCCGAGTCTTATTGACGTAAGGGA...
Embodiment 2
[0066] Example 2: Steps for generating a 43nt single-stranded product at 65°C using CP4 epsps of transgenic soybean GTS 40-3-2 as a template.
[0067] CP4 epsps template sequence:
[0068] TGGGGTTTATGGAAATTGGAATTGGGATTAAGGGTTTGTATCCCTTGTGCCATGTTGTTAATTTGTGCCATTCTTGAAAGATCTGCTAGAGTCAGCTTGTCAGCGTGTCTCTCCAAATGAAATGAACTTCCTTATATAGAGGAAGGGTCTTGCGAAGGATAGTGGGATTGTGCGTCATCCTTACGTCACTGGGAGATATCACATGGTTCCACTTGCTTTGAAGACTTACGTG
[0069] Complementary sequence of CP4 epsps template:
[0070] AGGAGCATCGTGGAAAAAGAAGACGTTCCAACCACGTCTTCAAAGCAAGTGGATTGATGTGATATCTCCACTGACGTAAGGGATGACGCACAATCCCACTATCCTTCGCAAGACCCTTCCTCTATATAAGGAAGTTCATTTCATTTGGAGAGGACACGCTGACAAGCTGACTACTAGCAGATCTTTCATAAGAATGGCACAAATTAACCACCATGGCACAAGGGATACAACCCAT.
[0071] Preparation of reagents before reaction: Add 50ul reaction reagents to the PCR tube: 10U Nb.BsrDI, 9.6U Bst DNAPolymerase, 2.5ul NEBuffer 3, 5ul Thermopol Reaction Buffer, 300uM dNTP, 200nM primers F1 and R1, 50nM primers BF and BR, 2.5ul template, 50nM nuc...
Embodiment 3
[0098] Example 3: Steps for generating a 30nt single-stranded product at 35°C using the 16S rDNA of citrus Huanglongbing HLB as a template.
[0099] 16S rDNA template sequence:
[0100] GGGTTGCCCCCATTGTCCAATATTCCCCACTGCTGCCTCCCGTAGGAGTCTGGGCCGTGTCTCACGTCCCAGTGTGGCTGATCGTCCCTCTCAGACCAGCTATAGATCGTAGCCTTGGTAGGCTCTTACCCTACCAACTAGCTAATCCAACGCAGG.
[0101] Complementary sequence of 16S rDNA template:
[0102] CCTGCGTTGGATTAGCTAGTTGGTAGGGTAAGAGCCTACCAAGGCTACGATCTATAGCTGGTCTGAGAGGACGATCAGCCACACTGGGACTGAGACACGGCCCAGACTCCTACGGGAGGCAGCAGTGGGGAATATTGGACAATGGGGGCAACCC.
[0103] Preparation of reagents before reaction: Add 50ul reaction reagents to the PCR tube: Add 50ul reaction reagents to the PCR tube: 10U Nt.AlwI, 5U phi29 DNA Polymerase, 2.5ul NEBuffer 3, 5ul Thermopol ReactionBuffer, 300uM dNTP, 200nM primer F1, 1 uM primer R1, 50 nM primers BF and BR, 2.5 ul template, 50 nM nucleic acid repressor. Incubate at 35°C for 10 minutes.
[0104] The sequences of primers and nucleic acid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com