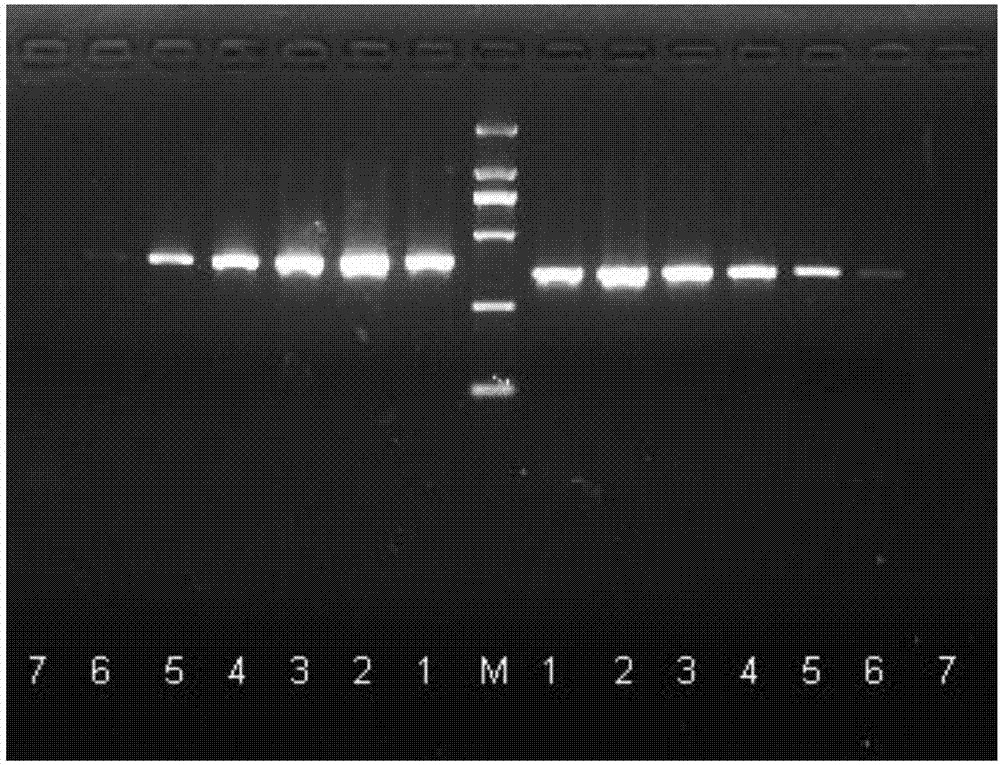
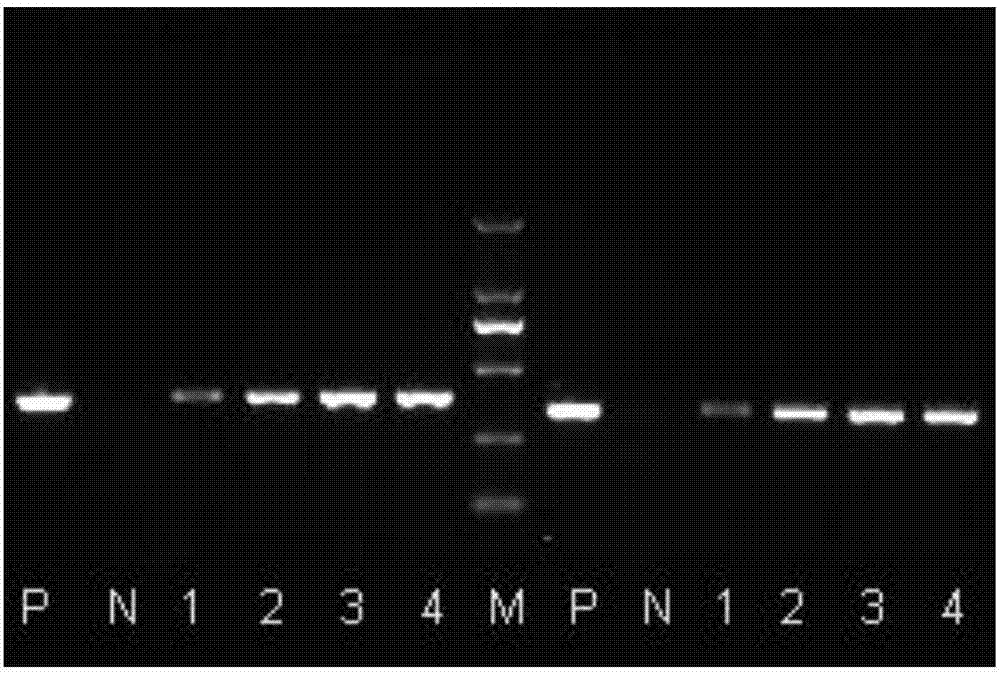
A crucian carp bafinivirus HB93, RT-PCR detection primers and applications of the bafinivirus and the primers
A RT-PCR, coronavirus technology, applied in applications, viruses, antiviral agents, etc., can solve the problem of no detection method, and achieve the effect of ensuring high specificity, strong specificity, and improving detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] A crucian carp coronavirus HB93, its isolation process is as follows:
[0041] 1) Take out and grind crucian carp liver, brain, spleen, kidney and other tissues carrying the virus, dilute the culture medium by 100 times, centrifuge at 2000rpm, take the supernatant and filter, and inoculate into grass carp gonad cells (Grass carp ovary cell line, GCO) The monolayer was cultured at 25°C for 7 days, resulting in cytopathic effect (CPE). It can be seen that the GCO cells shrink and become rounded, the refractive index increases, some cells begin to fall off, and the cell monolayer begins to rupture.
[0042] Medium: M199, 10% fetal bovine serum, pH 7.0-7.2
[0043] 2) Collect the diseased cells, and use transmission electron microscopy for morphological identification.
[0044] 3) The virus is collected, and its whole genome is sequenced, and the sequence is shown in SEQ ID NO.1.
[0045]Morphological characteristics of crucian carp coronavirus: The virus is rod-shaped, s...
Embodiment 2
[0047] Primers designed based on the difference between the full sequence of crucian carp coronavirus HB93 (shown in SEQ ID NO.1) and the full sequence of other viruses:
[0048] The present invention takes ordinary PCR as an example for illustration, and other primers designed based on the differential sequence, fluorescent quantitative primers, LAMP primers, digital PCR primers, etc., can also complete the detection of crucian carp coronavirus HB93.
[0049] A crucian carp coronavirus HB93 RT-PCR detection primer, including F1: 5'-CAAAGTAAACCCTGGAGA-3', R1: 5'-ATCAATGATGGTGCAGTT-3'; F2: 5'-TGAAACAATCCAAGAAGA-3', R2: 5'-ACAAGTTATAGCCGGTGT-3 '.
Embodiment 3
[0051] Utilize crucian carp coronavirus HB93RT-PCR to detect primers to the detection of crucian carp coronavirus HB93, the method comprises:
[0052] 1. Extraction of total RNA from crucian carp tissue or infected cells and synthesis of cDNA: using Extract the total RNA of the sample to be tested. Configure the reverse transcription reaction system: 3 μL of total RNA, 1 μL of Oligo(dT)18 primer, 8 μL of sterile double distilled water, mix well at 65°C for 5 minutes and immediately put it on ice; add 4 μl of 5×Reaction Buffer, 1 μl of RNase inhibitor, 2 μL of 5mM dN TPs , M-MLV reverse transcriptase 1μL was added to the reaction solution in ice bath, after mixing, 42°C for 1h, 95°C for 5min, and the obtained cDNA template was stored at -20°C for later use.
[0053] 2. Polymerase chain reaction amplification:
[0054] Use 25μl reaction system: Take 0.5μL of two pairs of primers F1, R1 or F2, R2 with a concentration of 40μM and add them to two PCR reaction tubes, then add 2μL...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com