Method for constructing DNA (deoxyribonucleic acid) sequencing library of to-be-detected genome and application thereof
A DNA sequencing and genome technology, applied in the biological field, can solve the problems of limited cell number, inability to obtain effective and high-resolution genome three-dimensional structure information, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
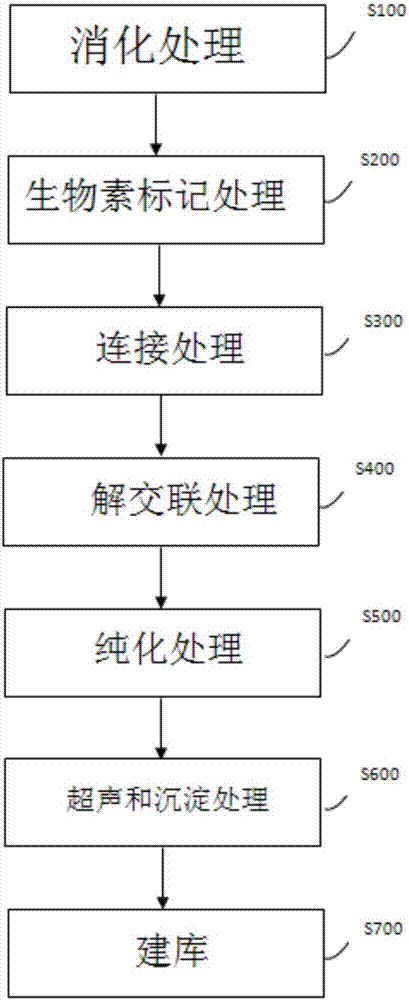
[0050] Example 1 Construction of DNA sequencing library of cell genome
[0051] 1.1 Reagent preparation
[0052] Lysis buffer
[0053] 10mM Tris-HCl., pH=7.4
[0054] 10mM NaCl
[0055] 0.5% NP-40
[0056] 0.1mM EDTA
[0057] 1X Proteinase Inhibitor
[0058] 2X biotin and streptavidin magnetic bead binding solution (2X Binding buffer)
[0059] 10mM Tris-HCl., pH=8.0
[0060] 2M NaCl
[0061] 1 mM EDTA
[0062] Tween wash buffer
[0063]5mM Tris-HCl., pH=8.0
[0064] 1M NaCl
[0065] 0.05%Tween
[0066] 0.5mM EDTA
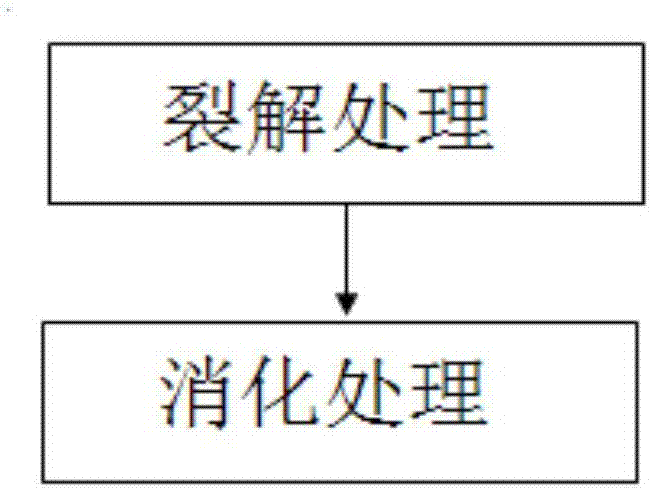
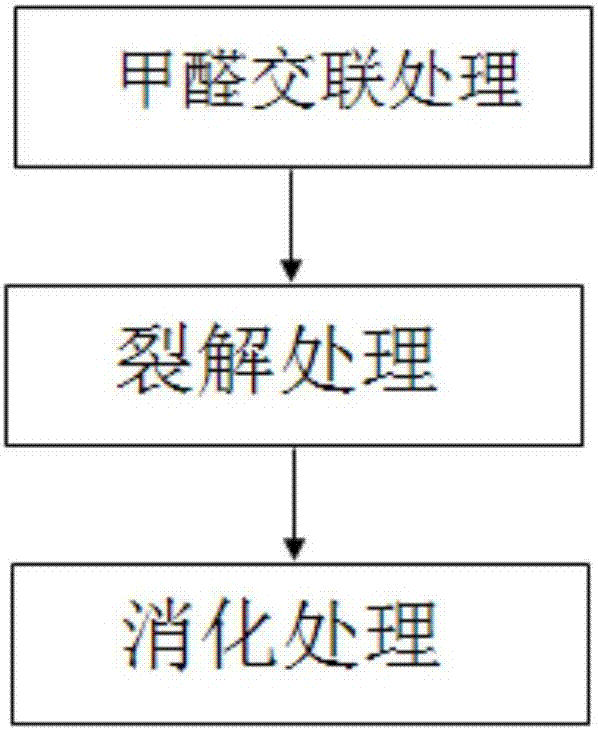
[0067] 1.2 Formaldehyde crosslinking
[0068] Transfer the collected samples to a freshly prepared PBS solution containing 1% formaldehyde under a stereoscope with a mouth pipette, fix at room temperature for 10 minutes, add 2.5M glycine solution to a final concentration of 0.2M, and let stand at room temperature for 10 minutes. Use a mouth pipette to transfer the sample into the PBS solution to wash once, then transfer to a PCR tube.
[0069] 1.3 Cl...
Embodiment 2
[0086] In order to compare the sisHi-C of the present application with the previous method (Rao et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatinlooping. Cell. 2014) in the effect of a small number of cells, the inventor used a mouth pipette to accurately Four groups of mouse embryonic stem cells were counted. The number of cells in each group was 500. Then, the inventors used sisHi-C and previous methods to build banks for two groups of cells (respectively labeled as repeated experiment 1 and repeated experiment 2). The concentration of DNA was measured before PCR, and the proportion of effective data was analyzed after library sequencing. Figure 8 The results of A show that before PCR, the DNA concentration of the two groups using the sisHi-C method is significantly higher than that of the two groups using the previous method; Figure 8 The results of B show that sisHi-C significantly reduces the loss of DNA. In the sequencing data...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com