Specific primers and screening method for EST-SSR (Express Sequence Tag-Simple Sequence Repeat) marker of torreya grandis transcriptome sequence
A technology of transcriptome sequence and specific primers, which is applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc. It can solve the problems of complex steps, high development costs, and no EST-SSR marker research reports.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
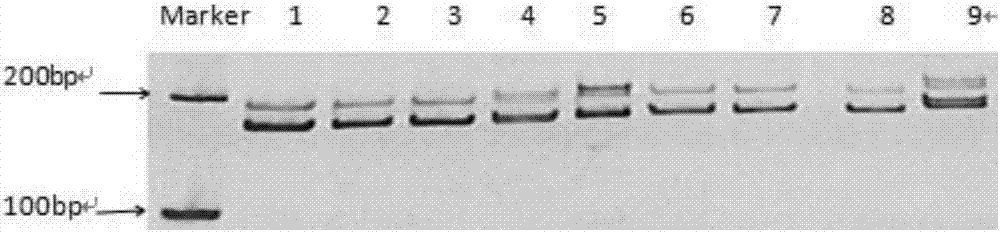
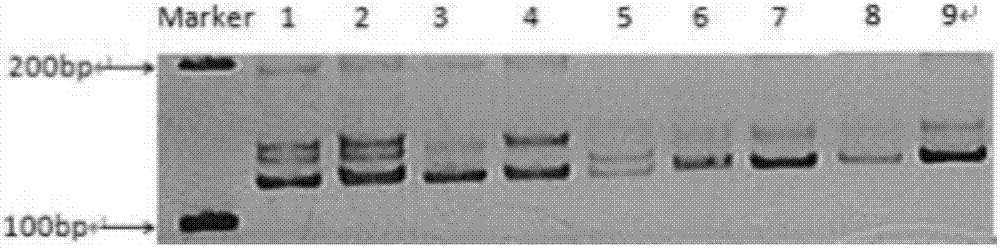
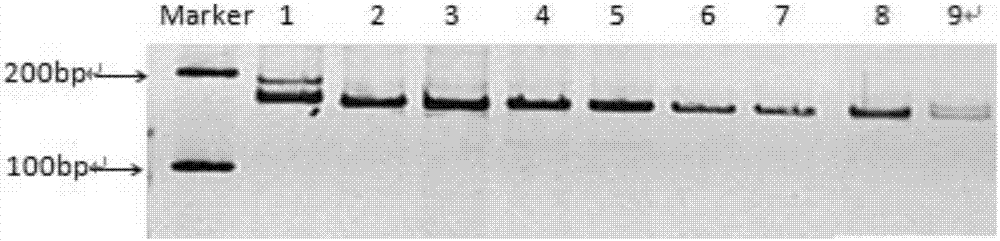
[0138] 1. Source of SSR loci and screening of EST-SSR sequences
[0139] The inventors used Trinity software to assemble the transcriptome data of the small torreya seed kernel transcriptome data and the round torreya seed kernel transcriptome data in the laboratory, and obtained the transcript sequence. A total of 246862 Transcripts and 142213 Unigenes were assembled. Use the MISA program (MIcroSAtellite identification tool, http: / / pgrc.ipk-gatersleben.de / misa / misa.html) to search for the SSR site of the Unigene above 1 kb obtained through screening. For the microsatellite fragments whose repetition times are 9, 8, 5, 5, and 5 times in sequence for repeating units of 2-6 bases, ESTs sequences containing microsatellite repeats are obtained. Using the primer design software Primer3.0 to design specific primers on the flanking sequences at both ends of the microsatellites.
[0140] 2. Design of EST-SSR labeled primers
[0141] In the flanking sequence of the microsatellite re...
Embodiment 2
[0150] 1. Extract the DNA of the Torreya tree
[0151] Specific steps are as follows:
[0152]Put CTAB in a 65°C water bath for 30 minutes in advance, take a fresh sample with an ice box, cut it into pieces and put it in a 2ml centrifuge tube (the sample volume is about 1 / 4 of the tube). Add a steel ball with a diameter of about 4mm into each centrifuge tube, soak it in liquid nitrogen for 5-7 minutes, fully pre-cool it, and grind it with a homogenizer. Add 800 μl of CTAB extract that has been water-bathed in advance to the centrifuge tube (add 1% β-mercaptoethanol to the CTAB extract), place the centrifuge tube in a 65°C water bath for 40 minutes, and shake it upside down once during the period. Centrifuge at 12000rpm at 4°C for 10min, take the supernatant, add an equal volume of 24:1 chloroform-isoamyl alcohol (chloroform:isoamyl alcohol=24:1), shake well, and let stand for 5-8min. 12000rpm, 4℃, centrifuge for 10min, take the supernatant, add an equal volume of 24:1, shake...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com