Method for fast identifying drug resistance of corynespora cassiicola on fluopyram and special primer pair
A technology for fluopyram and Corynespora multiprimum, applied in the field of molecular biology, can solve time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1. Discovery of succinate dehydrogenase B subunit gene SdhB and its corresponding protein mutation sites in Corynespora polymata
[0056] 1. Determination of susceptibility of Corynespora polymata to fluopyram
[0057] 5 sensitive strains HG15050755-3, HG15041321-2, HG15041414-2, HG14102405-2 and HG14102524-4; 4 resistant strains HG15050729-5, HG15050731, HG15050737 and HG15050825-6.
[0058] Potato dextrose agar medium (PDA): 200 g of potatoes, 20 g of glucose, 15 g of agar, distilled water to 1 L, sterilized at 121° C. for 30 min.
[0059] 1) Mix fluopyram with acetone to make 5×10 4 μg / ml stock solution. For the sensitivity determination of 5 strains of Corynespora polymaine sensitive strains and 4 strains of Corynespora polymaine resistant strains, fluopyram was diluted to 10 μg / mL, 100 μg / mL, 500 μg / mL, 1000 μg / mL, 5000 μg / mL, 10000μg / mL and 25000μg / mL concentration gradient.
[0060] 2) In order from low to high, use a pipette to draw 135 μL of the dr...
Embodiment 2
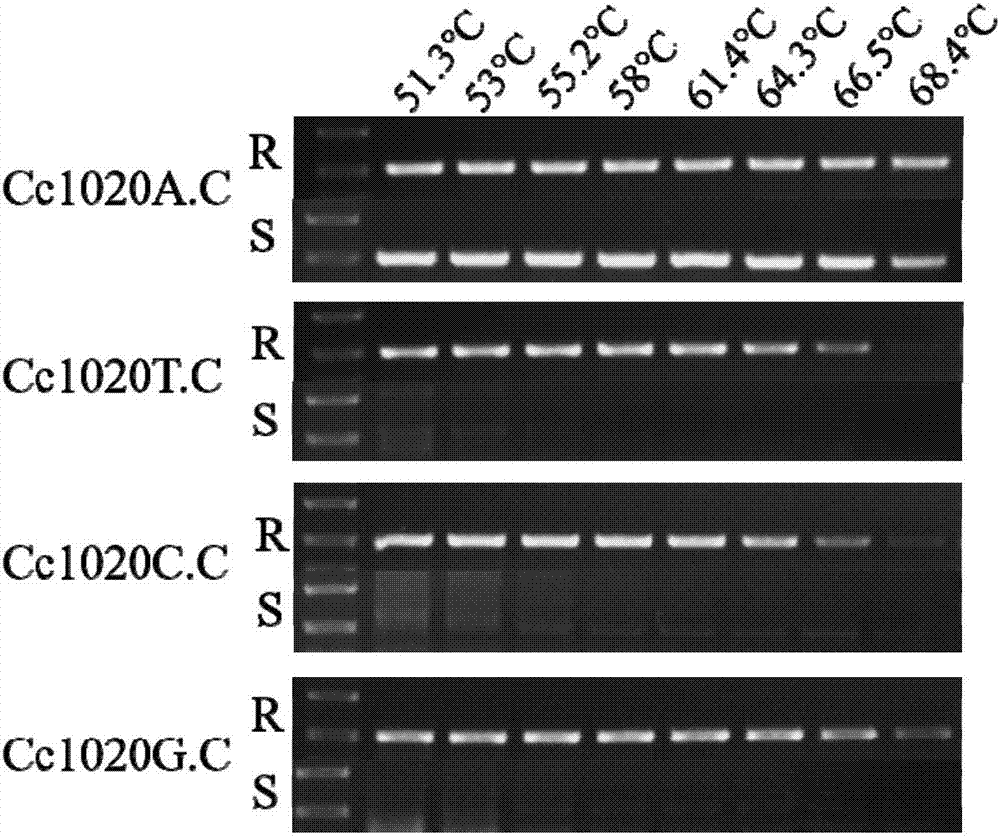
[0083] Embodiment 2, the method for detecting the mutation site A1020G in Corynespora polymata
[0084] 1. Primer design, screening and amplification methods
[0085] The strains are the strains HG15050729-5, HG15050825-6, HG14102524-4, HG14102405-2, HG15041321-2, HG15041414-2, HG15050755-3 used in Example 1.
[0086] AS-PCR primers were designed according to the sequence of the mutant HG15050729-5 succinate dehydrogenase B subunit gene SdhB. The primer pair Cc1020F-Cc1020A / T / C / G.C.R was used to detect the mutation site of A1020G. Wherein, the last base of the 3'-end of the reverse primer Cc1020A.C.R is the mutated base, and the forward primer is Cc1020F (Table 2). In order to increase the specificity of the primers, mismatched bases were introduced into the 2nd and 3rd bases at the 3'-end of the reverse primer (Table 2, primers Cc1020T / C / G.C.R).
[0087] Table 2 The PCR primers used in the detection of Corynespora polymorpha strains resistant to fluopyram
[0088]
[0...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com