Micromolecule for activating LRIG1 expression in brain glioma U251 cells and screening method and application of micromolecule
A technology for brain glioma and screening methods, applied in the direction of DNA/RNA fragments, organic active ingredients, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
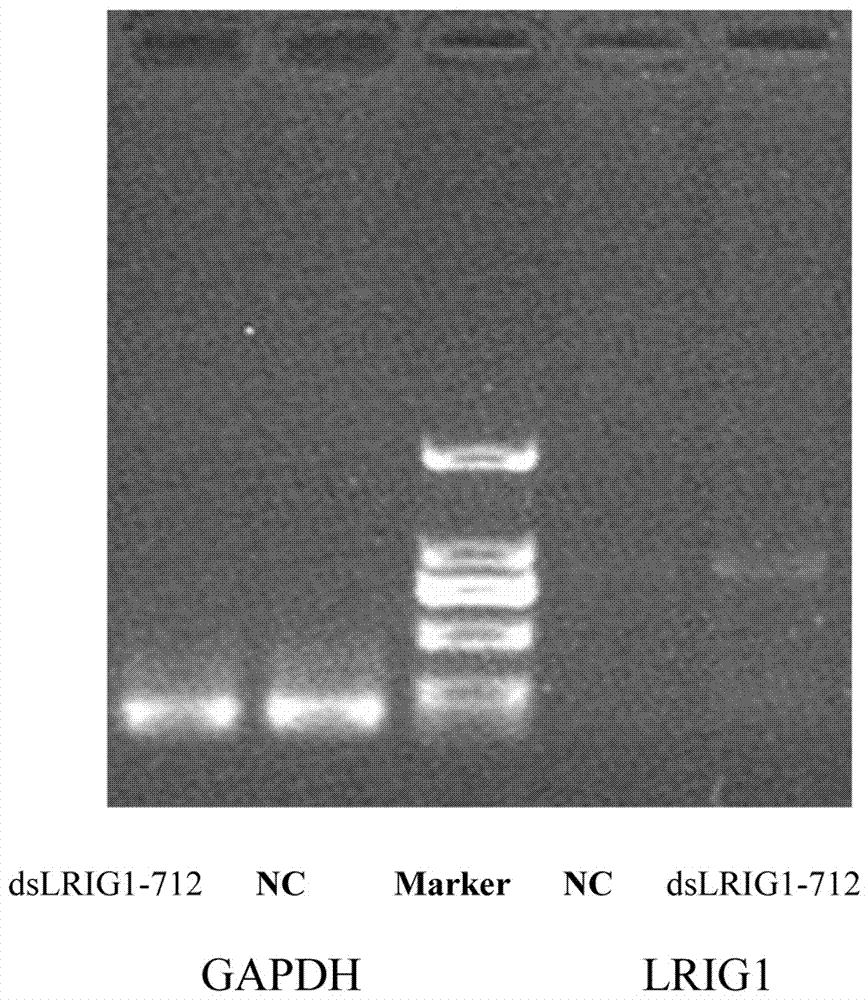
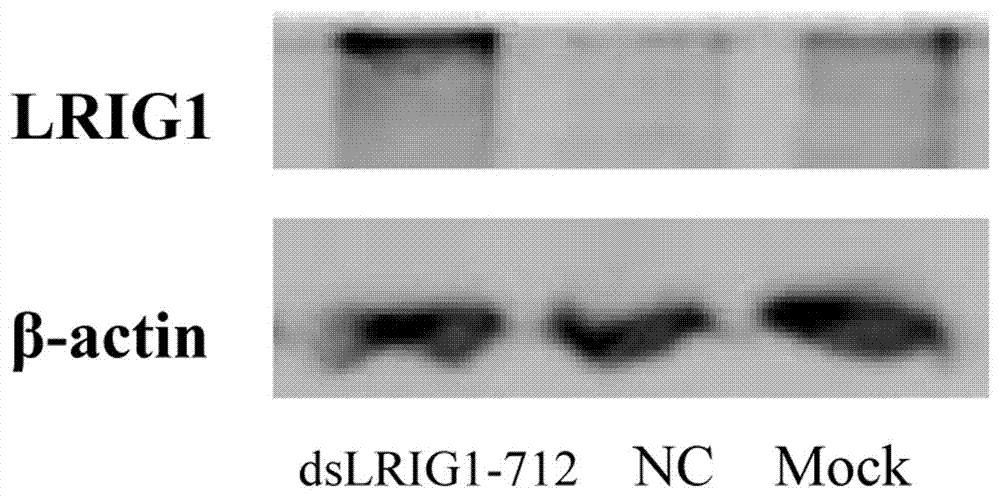
[0022] dsRNA Design dsLRIG1-712 is a dsRNA molecule with a length of 21 nucleotide base pairs designed for the relative transcription start point -712 of the LRIG1 gene promoter region and complementary to the promoter. The control dsRNA (dsControl) is non-homologous to the known human genome sequence, and is a dsRNA sequence with the same length of 21 nucleotide base pairs.
[0023] Use the Genebank database to find the sequence of the promoter region of the LRIG1 gene, and use the -712 sequence upstream of the promoter as a template for designing dsRNA. The specific sequence is: dsLRIG1-712: Sense: UUU CCC ACC CAA GGU GUG A[dT][dT]; Antisense: UCA CAC CUU GGG UGG GAA A[dT][dT].
[0024] Cell culture and transfection U251 cells were grown in DMEM medium containing 10% fetal bovine serum at 37°C, 5% CO 2 Incubator cultivation. The experiment was divided into three groups: blank group, control group (transfected with dsControl) and experimental group (transfected with candida...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com