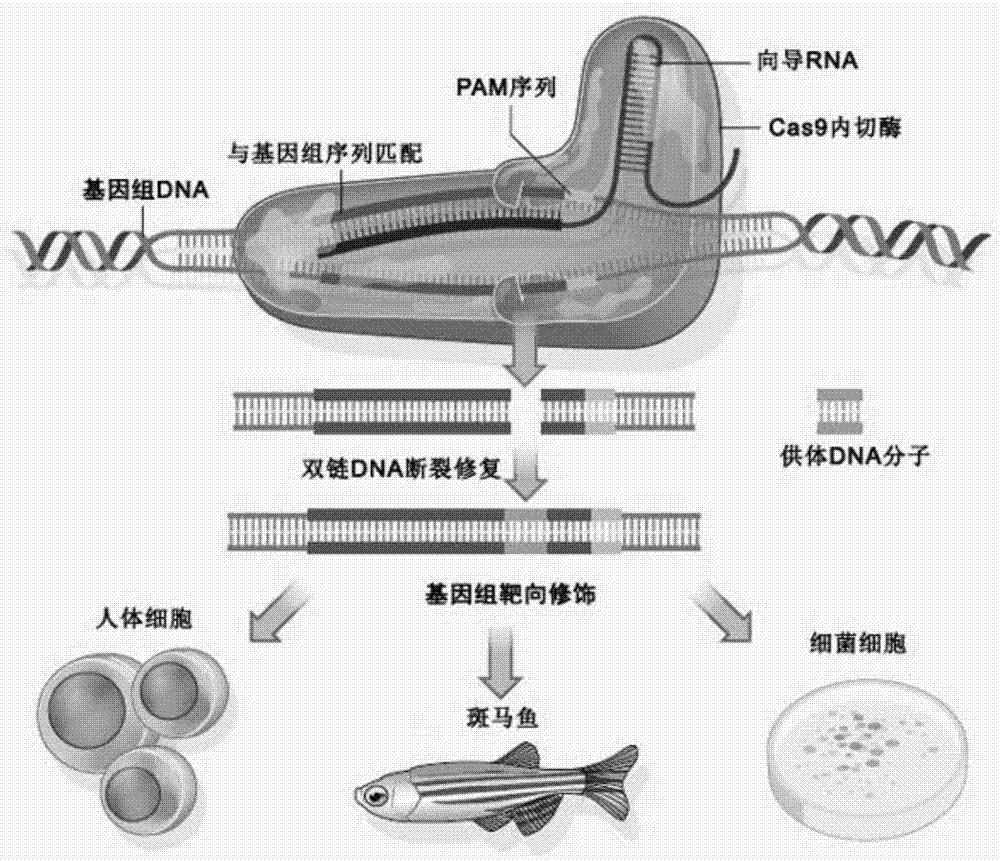
Method used for specifically increasing of CRISPR-CAS system gene editing efficiency
A gene editing and efficient technology, applied to other methods of inserting foreign genetic materials, genetic engineering, biochemical equipment and methods, etc., can solve the problems of low homologous recombination probability and affect precise editing, etc., to improve gene editing efficiency, The effect of improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1, cloning the potentiating protein CREnhancer1.0 and constructing the vector
[0032] The CREnhancer1.0 gene was cloned, and the gene sequence described in SEQ ID NO: 1 was obtained by the method of whole gene synthesis. Using this sequence as a template, according to the sequences of the upstream and downstream primers, they were 5'-ATGCAGGAGAACCTGGCCCCCTG-3', 5 '-CAGGCAGCTCACGCTCCTCTCG-3', primers and whole genome were synthesized by Shanghai Sangong Co., Ltd. The target gene fragment of CREnhancer1.0 gene was amplified by PCR reaction. The amplification reaction system was as follows: 95°C, 40s, 57°C, 1min, 72°C, 1min, 72°C, 10min, cycled 35 times, and the PCR product was produced by Shanghai Shenggong Co., Ltd. Sequencing was performed, and the binding was a complete match to SEQ ID NO:1 by sequencing. Subsequently, the target gene amplified by PCR was connected to the empty vector lentiviral vector pHIV-CS-CDF-CG-PRE, and the recombinant lentiviral vector...
Embodiment 2
[0033] Example 2 Application Analysis of CRISPR / Cas9 in Bone Marrow Mesenchymal Stem Cells
[0034] CRISPR / Cas9 editing vector based on pBGN plasmid containing BSD-fsEGFP fusion gene
[0035] (1) BSD-fsEGFP fusion gene: use conventional PCR to amplify the known BSD gene, 5'-PCR primers with HindIII sites, and 3'-PCR primers to introduce I-SceI and EcoRI sites. Insert the PCR product (BSD) into the HindIII and EcoRI positions between the CMV driver and the EGFP coding region in the EGFP plasmid (the EGFP nucleotide sequence is a sequence known in the art, such as shown in sequence 1 and sequence 2 in CN105647968A) Point, generate the plasmid pBGN containing the BSD-fsEGFP fusion gene, the nucleotide sequence of the BSD-fsEGFP fusion gene is as shown in sequence 3 and sequence 4 in CN105647968A). The fusion gene is driven by CMV driver or PGK driver, but EGFP is inactive due to frameshift, so it is called fsEGFP.
[0036] 5'-PCR primers are
[0037] CTCAAGCTTAACTAAACCATGGCCAA...
Embodiment 3 Embodiment 2
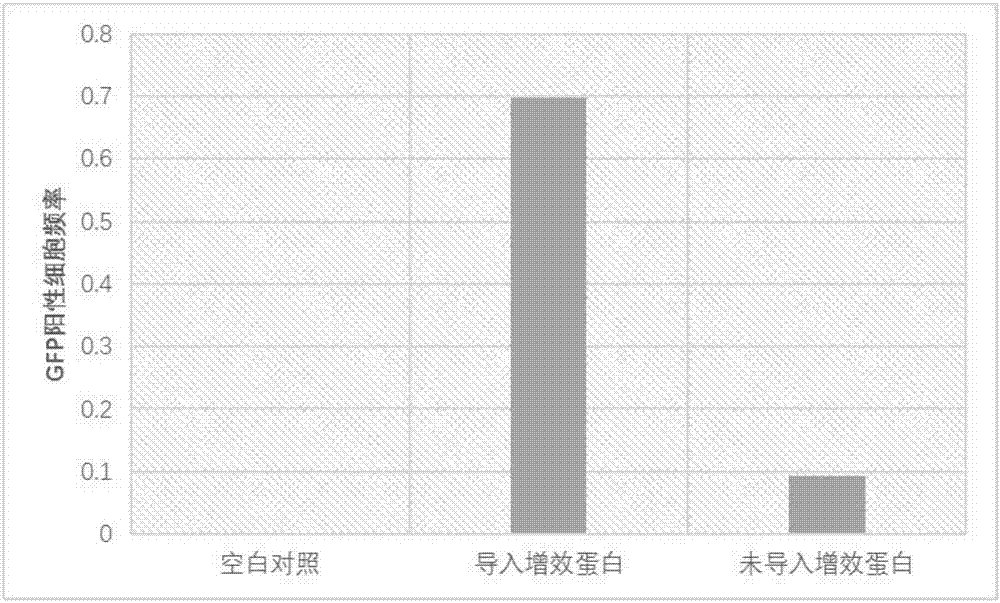
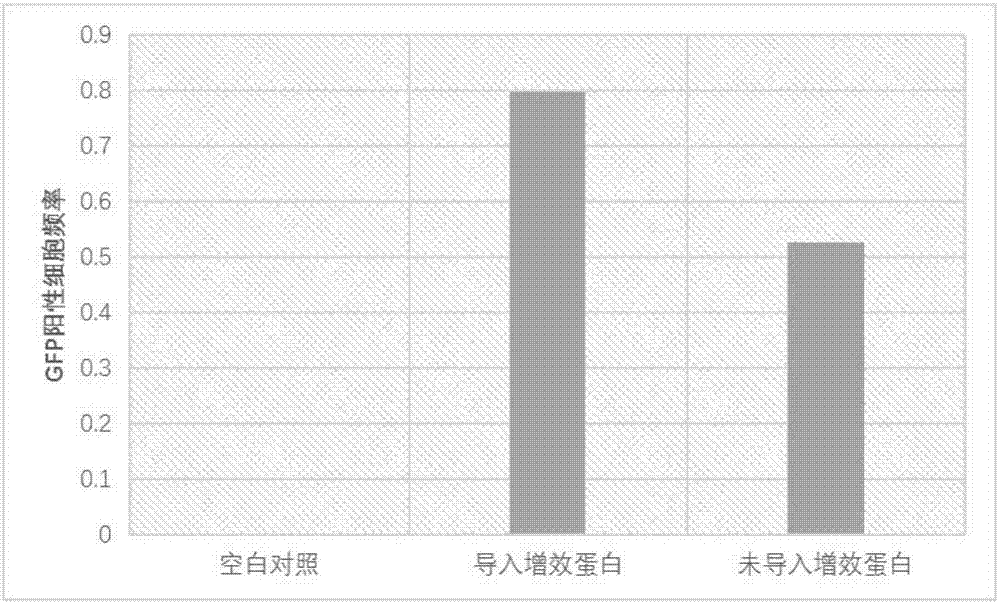
[0044] Example 3 Efficiency Analysis of the System of Example 2 in Human U2OS Cells
[0045] The system prepared according to Example 2 was transformed into human U2OS cells, and the human U2OS cells were respectively introduced or not introduced with the synergistic protein described in Example 1 as a control. HPRT-inactivated cells will develop resistance to 6-mercaptoguanine (6-TG) due to frameshifts or mutations caused by repair in human U2OS cells. After 4 weeks of screening using the known 6-TG screening method, 6-TG resistant cell clones will form, and the ratio of the formation frequency to the cell clone formation frequency without 6-TG selection represents true 6-TG resistant cells Clonogenic efficiency. This efficiency represents the efficiency with which the endogenous HPRT gene is edited. The result is as image 3 As shown, in U2OS cells, the endogenous HPRT gene editing efficiency mediated by the intracellular sgRNA without the synergist protein is 52.6%, and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com