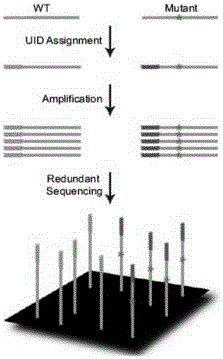
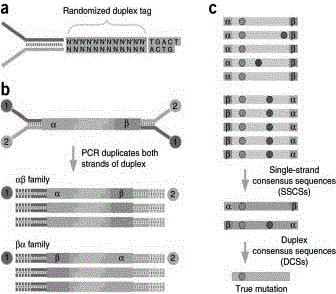
Constructing method of amplification sublibrary for precise sequencing of ctDNA
An amplicon library and construction method technology, applied in DNA preparation, recombinant DNA technology, chemical library and other directions, can solve problems such as cost increase, and achieve the effects of cost saving, improving sensitivity and efficiency, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
[0109] S01: Random ligation of cfDNA followed by random fragmentation and addition of adapters to build a library;
[0110] After extracting and purifying cfDNA, use Agilent 2100 or similar instruments to check the quality of cfDNA. The main peak of qualified samples should be distributed around 166bp. Take 10ng of cfDNA that has passed the quality inspection, and use T4 ligase to ligate for 5 hours, then use magnetic beads to purify the cfDNA, and dissolve it in 8 μL ddH2O; the ligation effect is judged by the distribution of fragment sizes.
[0111] Take 8ng of the above randomly linked cfDNA, add Tn5 transposome fragmentation system (5*lm lysis buffer 6μL, tn5 transposon 11μL, 10ng linked cfDNA, ddH2O supplemented to 30ul), react at 60°C for 10min, then The fragmented product was purified using magnetic beads and the purified product was dissolved in 35 μL ddHO.
[0112] Before random fragmentation of the cfDNA and addition of adapters, preparation of transposomes was also...
specific Embodiment 2
[0113] S01: Random ligation of cfDNA followed by random fragmentation and addition of adapters to build a library;
[0114] After extracting and purifying cfDNA, use Agilent 2100 or similar instruments to check the quality of cfDNA. The main peak of qualified samples should be distributed around 166bp. Take 30ng of cfDNA that has passed the quality inspection, and use T4 ligase to ligate for 3 hours, then use magnetic beads to purify the cfDNA, and dissolve it in 12 μL ddH2O; the ligation effect is judged by the distribution of fragment sizes.
[0115] Take 12ng of the above randomly linked cfDNA, add Tn5 transposome fragmentation system (5*lm lysis buffer 6μL, tn5 transposon 11μL, 10ng linked cfDNA, ddH2O supplemented to 30ul), react at 50°C for 5min, then The fragmented product was purified using magnetic beads and the purified product was dissolved in 45 μL ddHO.
[0116] Before the random fragmentation of the cfDNA and the addition of adapters, the preparation of transpos...
specific Embodiment 3
[0117] S01: Random ligation of cfDNA followed by random fragmentation and addition of adapters to build a library;
[0118] After extracting and purifying cfDNA, use Agilent 2100 or similar instruments to check the quality of cfDNA. The main peak of qualified samples should be distributed around 166bp. Take 20ng of cfDNA that has passed the quality inspection, and use T4 ligase to ligate for 4 hours, then use magnetic beads to purify the cfDNA, and dissolve it in 10 μL ddH2O; the ligation effect is judged by the distribution of fragment sizes.
[0119] Take 10ng of the above randomly linked cfDNA, add it to the Tn5 transposome fragmentation system (5*lm lysis buffer 6μL, tn5 transposon 11μL, 10ng linked cfDNA, add ddH2O to 30ul), react at 55°C for 8min, and then The fragmented product was purified using magnetic beads and the purified product was dissolved in 40 μL ddHO.
[0120] Before the random fragmentation of the cfDNA and the addition of adapters, the preparation of tra...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com