Corn lncRNA screening and classifying method based on ARM processor
An ARM processor, screening and classification technology, applied in the field of bioinformatics research, can solve the problems of inability to calculate within a reasonable time, without any optimization, and high time complexity, and achieve easy batch processing of data, high accuracy, and high code. The effect of density
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0025] The present invention will be further described in detail below in conjunction with the accompanying drawings, so that those skilled in the art can implement it with reference to the description.
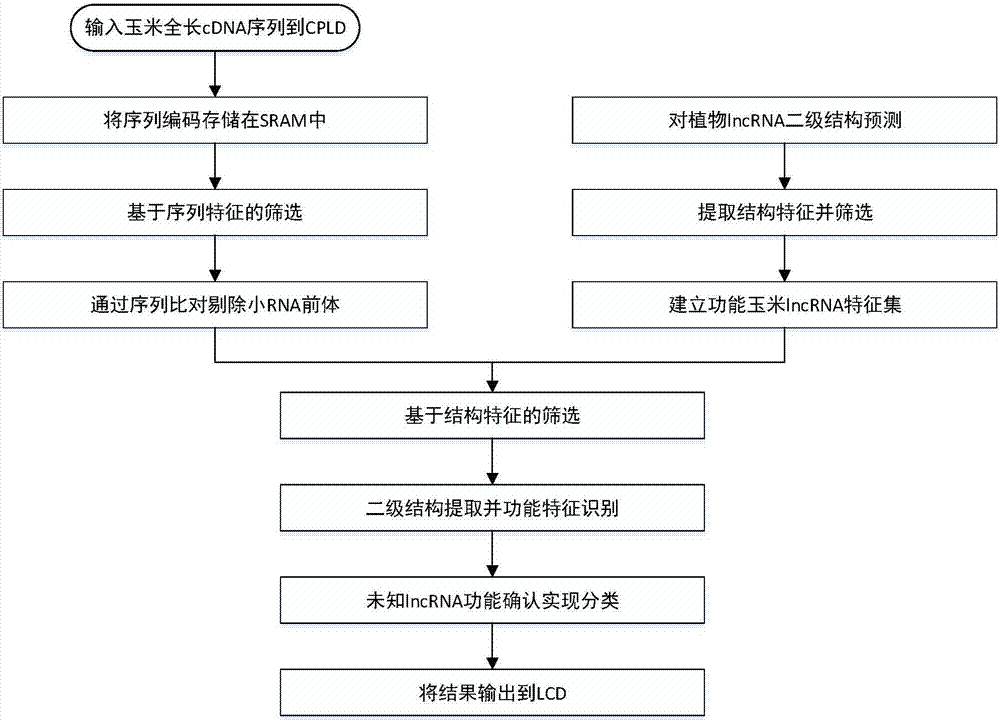
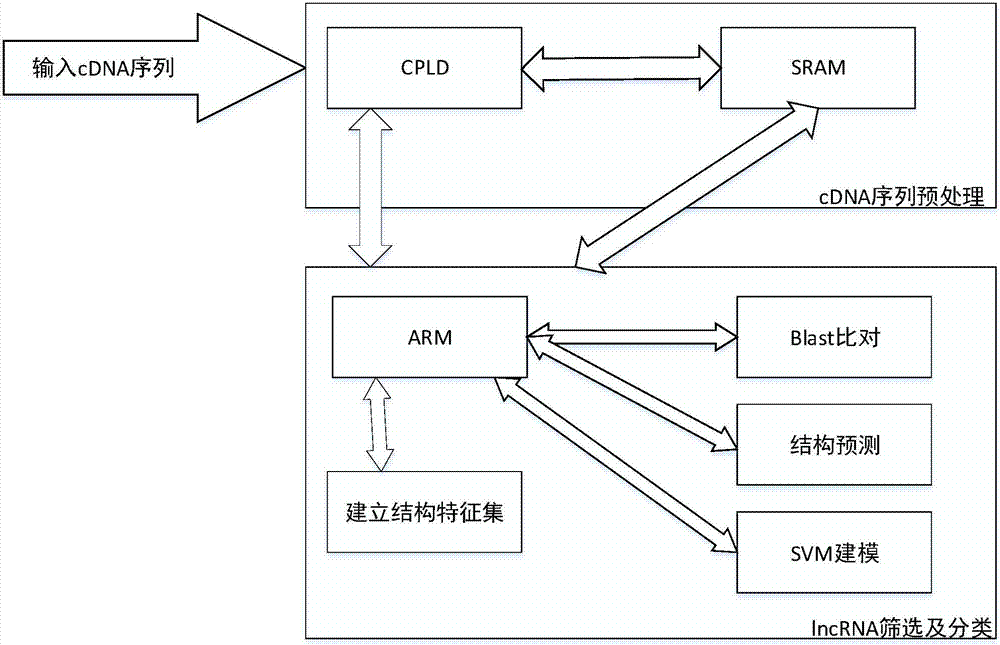
[0026] Such as figure 1 , figure 2 As shown, the existing lncRNA screening and classification methods first collect high-throughput lncRNA data according to traditional design rules, and then screen and classify them. The collection of lncRNA needs to be carried out with biological experimental equipment. The current method is not convenient to combine with high-throughput data collection equipment, etc., and the labor cost is high, the speed is slow, and the efficiency is low. Therefore, it is urgent to solidify the lncRNA classification method to high-throughput data In the collecting instrument, optimize lncRNA screening classification; Therefore, the invention provides a kind of corn lncRNA screening classification method based on ARM processor, the required hardware eq...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com