Detection method of coxsackievirus A10 in mixed infection of hand-foot-mouth disease
A technology of mixed infection and hand, foot and mouth disease, applied in the biological field, can solve the problems of huge cost and increase cost, and achieve the effect of reducing cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
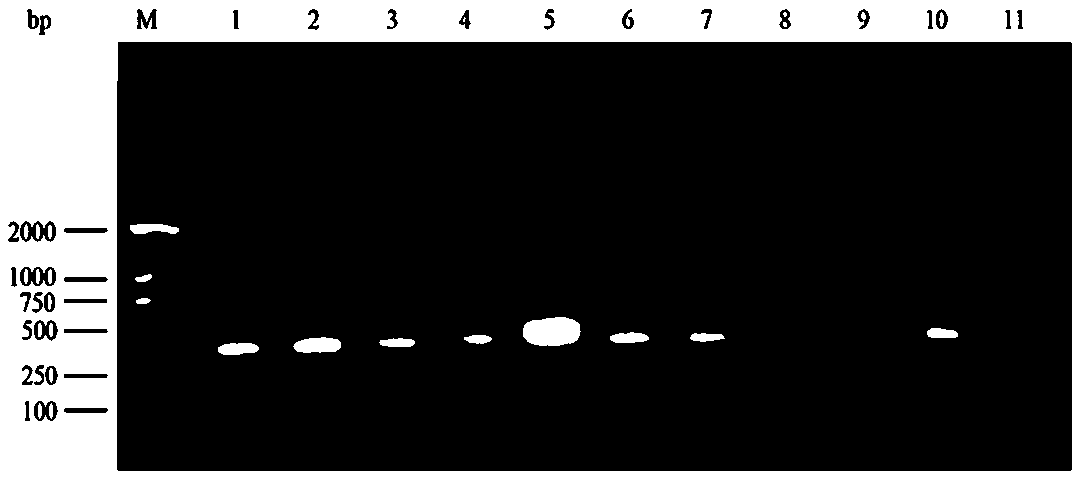
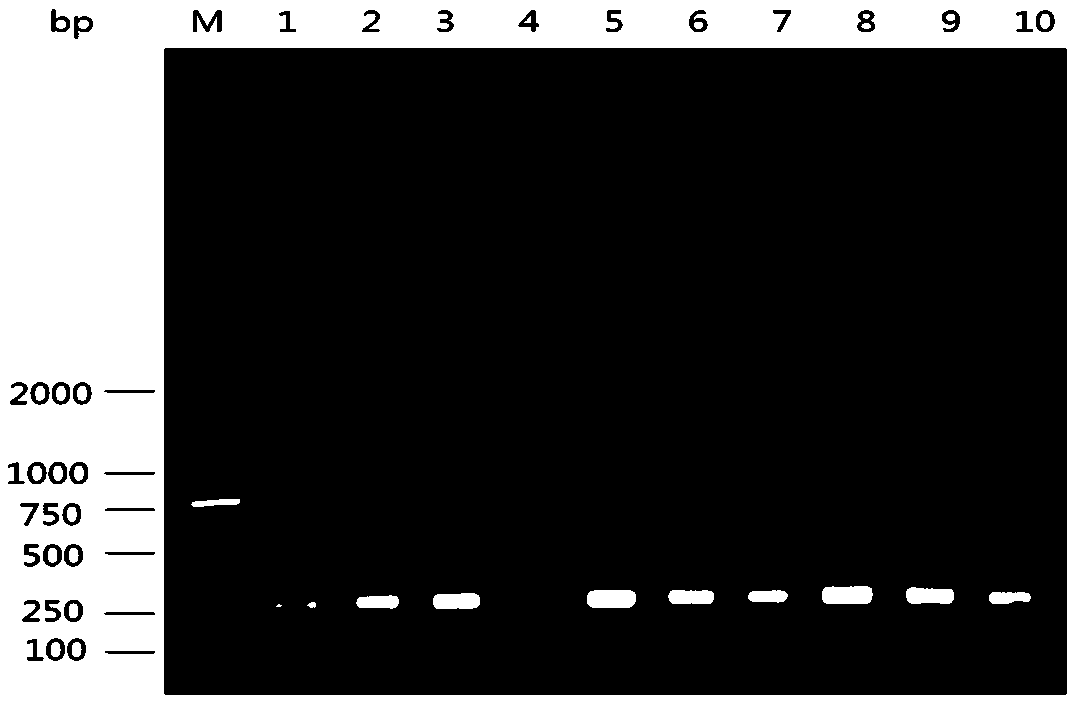
[0020] 1. Sample processing
[0021] Take 0.5 g of feces, add 2.5 mL of PBS to shake and mix well, centrifuge at 3000 r / min for 30 min, take the supernatant, and store it at -20 °C;
[0022] 2. Viral RNA extraction
[0023] The extraction of viral RNA was performed according to the instructions of the kit (Qiamp viral RNA mini Kit), as follows: Take 140 μL of supernatant or cerebrospinal fluid and add 560 AVL, mix it upside down and place it at room temperature for 10 min; add 560 μL of absolute ethanol, mix it upside down; mix 630 μL of the above solution Add to the adsorption column, centrifuge at 8000rpm for 1min, add 500μLAW1, centrifuge at 8000rpm for 1min, pour off the waste liquid in the collection tube; add 500μLAW2, centrifuge at 14000rpm for 3 minutes, pour out the waste liquid in the collection tube; put the adsorption column Put it into an empty collection tube, centrifuge at 14000rpm for 1min; put the adsorption column into a new centrifuge tube, add elution buff...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com