Primer set, reagent and kit for detecting methylation of RPRM gene and PRDM5 gene and use thereof
A technology of methylation and primer set, applied in the biological field, can solve the problems such as inability to obtain pathological diagnosis, and achieve the effect of easy mastery, strong specificity and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] The composition of embodiment 1 kit and the processing of sample to be tested
[0089] 1. The kit for simultaneously detecting methylation of RPRM gene and PRDM5 gene provided by the present invention includes a DNA processing kit and a methylation gene detection kit. Among them, the composition of the DNA processing kit is shown in Table 1, and the composition of the methylated gene detection kit is shown in Table 2.
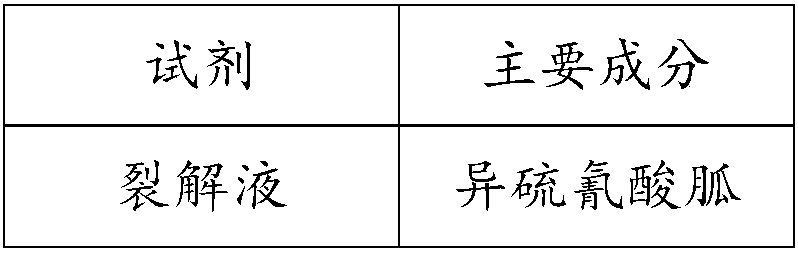
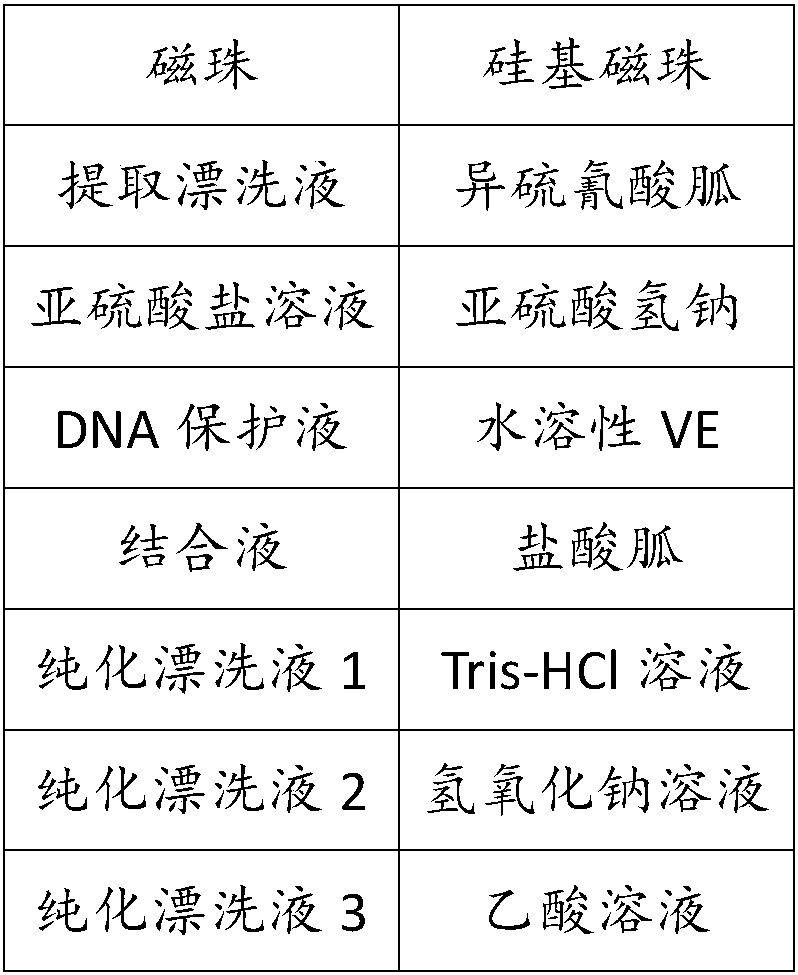
[0090] Table 1 DNA processing kit
[0091]
[0092]
[0093] Table 2 Methylation gene detection kit
[0094] Reagent
main ingredient
PCR Mix
dNTPs, primers, buffer salts
Taq Enzyme Mix
Taq enzyme, probe, buffer salt
TE buffer, unmethylated human genomic DNA
positive control substance
TE buffer, methylated human genomic DNA
[0095] 2. Instruments and equipment: The instruments and equipment required for testing are as follows:
[0096]Vortex shaker, rotary mixer, 15mL ...
Embodiment 2
[0123] Example 2 Detection of RPRM and PRDM5 methylated genes
[0124] 1) According to the reaction system in Table 3, prepare the PCR pre-reaction solution, mix it and distribute it into PCR tubes, add the sample to be tested, the reference substance and purified water as templates, and the volume of each reaction is 20 μL;
[0125] Table 3 PCR reaction system
[0126]
[0127] 2) Perform fluorescent quantitative PCR reaction according to the reaction conditions in Table 4.
[0128] Table 4 reaction conditions
[0129]
[0130] 3) After the sample is put on the machine, save the experiment name, run the PCR reaction, and write the sample layout and sample name according to the experiment layout in time.
Embodiment 3
[0131] Embodiment 3PCR result analysis
[0132] 1. in Select "Analysis" in the 480 basic software module column.
[0133] 2. Select the "Abs Quant / Fit Points" analysis mode.
[0134] 3. Select "Filter Comb 465-510".
[0135] 4. Select the "Cycle Range" window, set "First Cycle" to "1"; "Last Cycle" to "50".
[0136] 5. In the "Cycle Range" window, set the background to "5-22", click the blue "Background" button, set the "Min Offset" to "4", and set the "Max Offset" to "twenty one".
[0137] 6. Select the "Noise Band" window, set "Noise Band" to "Noise Band (Fluoresc)", and manually change the value of "Noise Band" to "2.0".
[0138] Note: If the background value is higher than 2.0, the baseline can be adjusted according to the actual situation, that is, the value of "Noise Band", until the amplification curve without obvious exponential growth period should be negative.
[0139] 7. Select the "Analysis" window, "Threshold(Auto)" will automatically adjust the "Threshold"...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com