Method for detecting DNA glycosylase activity by alpha-hemolysin nanopores
A technology of glycosylase activity and glycosylase, which is applied in the field of biochemical analysis, can solve the problems of complex operation and low sensitivity, and achieve the effects of high sensitivity, improved sensitivity and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
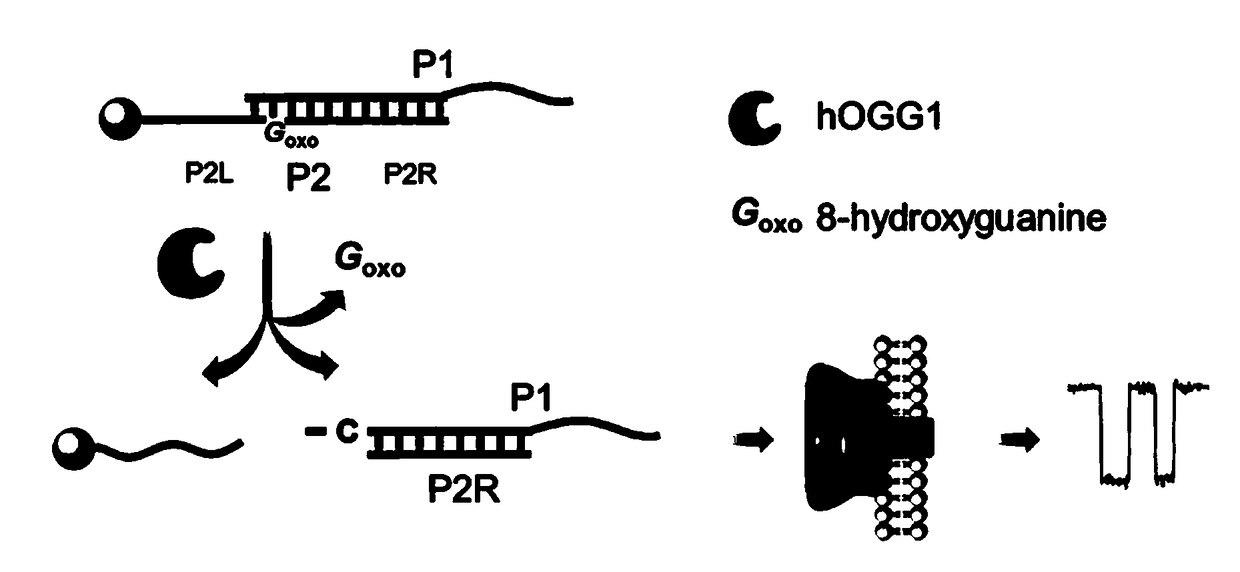
[0037] (1) Preparation of double-stranded DNA substrate probe
[0038] 1) Prepare a double-stranded DNA substrate probe containing the damaged base 8-oxoguanine (8-oxoG): DNA strand P1 (40 μL, 10 μM) and DNA strand P2 (40 μL, 10 μM) in 20 μL buffer (150 mM NaCl , 20mM Tris-HCl, pH 7.9), incubated at 95°C for 5 minutes and gradually cooled to room temperature to obtain double-stranded DNA with a concentration of 4μM, which was named P1 / P2. The sequence of P1 is 5'-ACGACAGAGTAGGATTCTCGACC 30 -3', the sequence of P2 is 5'- / oxoG / TCGTT 20 -biotin-3', where oxoG represents the damaged base 8-oxoguanine, the bolded base is named as the P2R sequence, the italicized base is named as the P2L sequence, and the 3' end of P2 is modified with a biotin molecule.
[0039] 2) Double-stranded DNA P1 / P2 was immobilized on the surface of streptavidin magnetic beads (MB): Firstly, the magnetic beads (50 μL, 10mg / mL) and wash three times, then mix 25μL double-stranded DNA, 25μL ultrapure wate...
Embodiment 2
[0044] Experimental Feasibility Verification
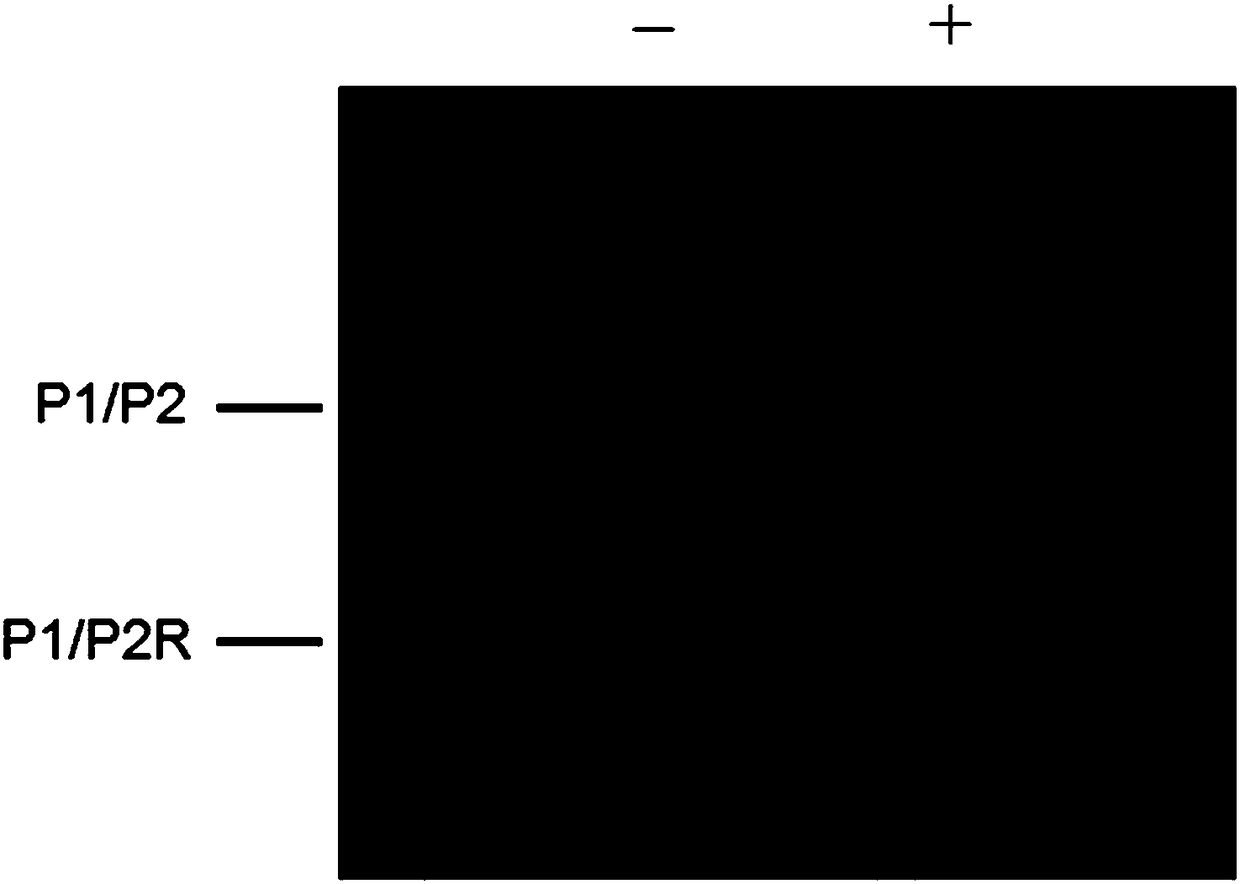
[0045] In order to verify the feasibility of DNA glycosylase hOGG1 excision of damaged bases in vitro, the reaction products were initially analyzed by non-denaturing polyacrylamide gel (PAGE) electrophoresis, and the results were as follows figure 2 shown. Since magnetic beads affect electrophoretic detection, double-stranded DNA (named P1 / P2) without magnetic beads was used as the reaction substrate. from figure 2 It can be seen that there is only one band when there is only double-stranded DNA substrate P1 / P2 and no DNA glycosylase hOGG1. When DNA substrate P1 / P2 and DNA glycosylase hOGG1 exist at the same time, two bands can be observed, one is a double-stranded DNA substrate, and the other is significantly smaller than the double-stranded DNA substrate, indicating that DNA glycosylase hOGG1 Specifically recognize and excise the damaged base, and generate a new, shortened partially complementary DNA, which is speculated t...
Embodiment 3
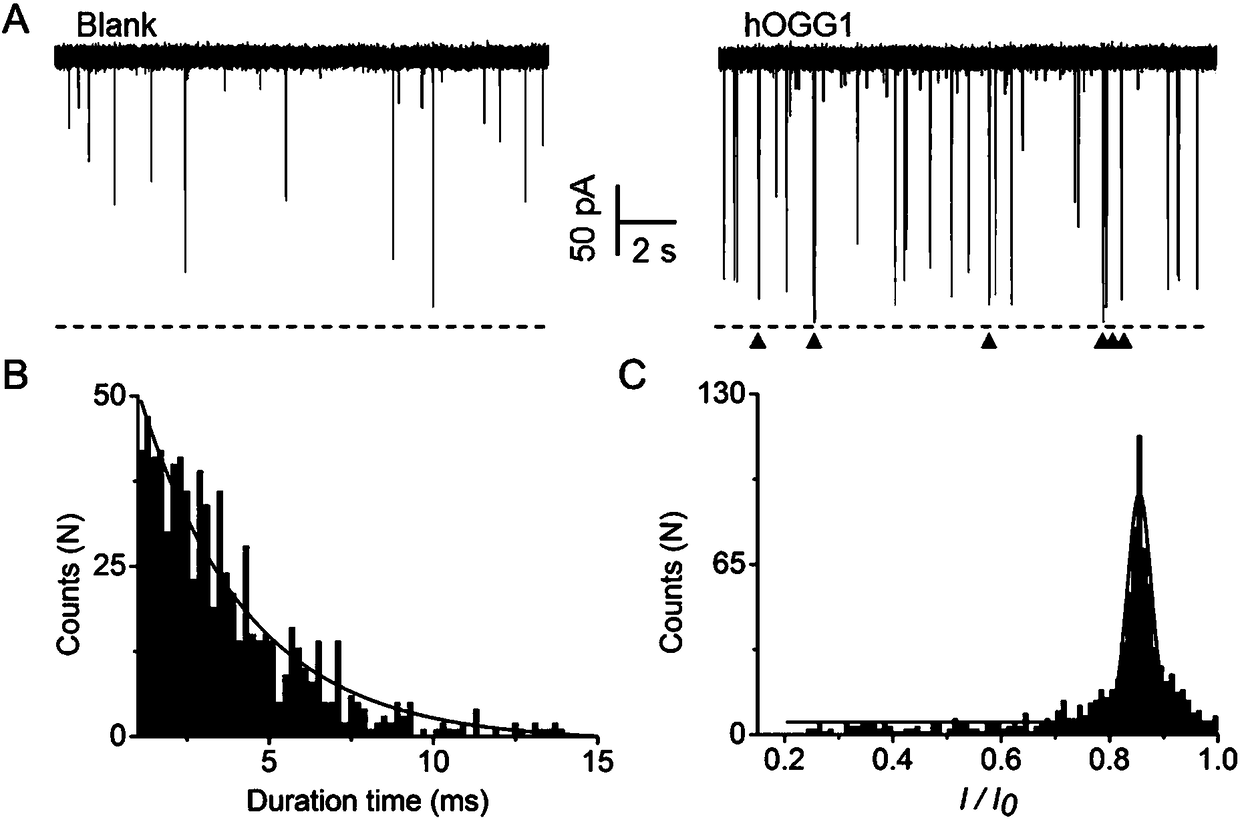
[0047] The nanopore of the present invention detects hOGG1 activity such as image 3 shown. (A) Nanopore detection map with and without hOGG1. Blank means the sample without hOGG1, only the background signal appears in the nanopore detection, which is speculated to be caused by the random collision between the reaction mixture and the opening of the nanopore; hOGG1 means the blocking signal when the DNA glycosylase hOGG1 exists, and the red triangle means the characteristic sex blocking signal. (B) Statistical analysis of the blocking time of the signal. (C) Statistical analysis of the degree of signal blockade. According to the statistical analysis results, the signal with a blocking time of 1-15 ms and a blocking degree of more than 80% is a characteristic blocking signal produced by the product P1 / P2R, which can be used as the basis for the existence of hOGG1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com