Efficient DNA editing method
A DNA sequence and editing technology, applied in the field of efficient DNA editing, can solve problems such as inability to cut, and achieve the effect of improving the ability of mismatched bases
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] An efficient DNA editing method, knock out the co-GFAP gene containing the co-GFAP plasmid in 293T cells. Specifically through the following methods:
[0019] 1) According to the sequence of SEQ ID NO.1, express and purify the fusion protein in prokaryote;
[0020] 2) Add the oligonucleotides of Table 1 to the above purified protein. The mixture is then added to the cultured cells.
[0021] 3) The co-GFAP gene can be knocked out by incubating for 72 hours again.
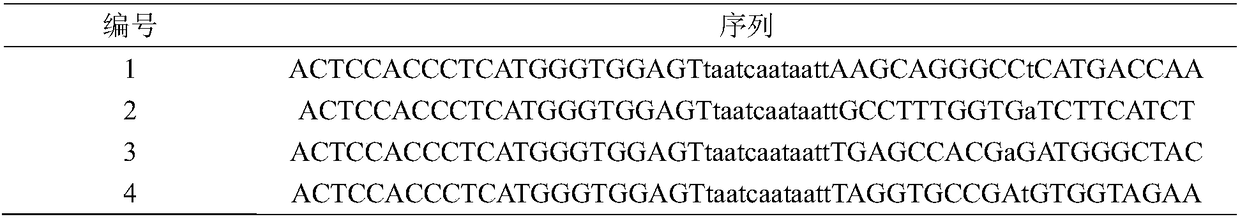
[0022] Table 1 Oligonucleotide sequence
[0023]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com