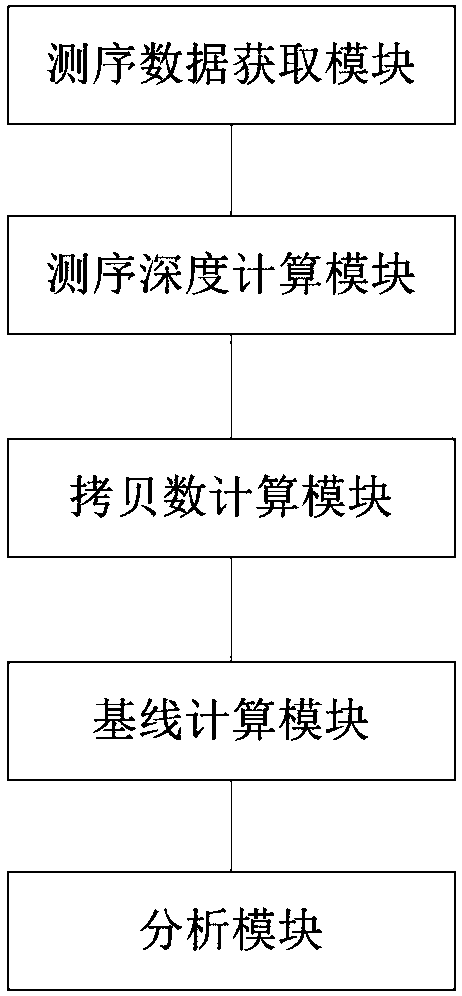
Detection methods and devices of copy number variations (CNVs) and computer-readable medium
A technology for copy number variation and detection method, applied in the field of copy number variation detection based on high-throughput sequencing data, can solve problems such as unstable calculation results, and achieve the effect of avoiding large fluctuation of copy number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
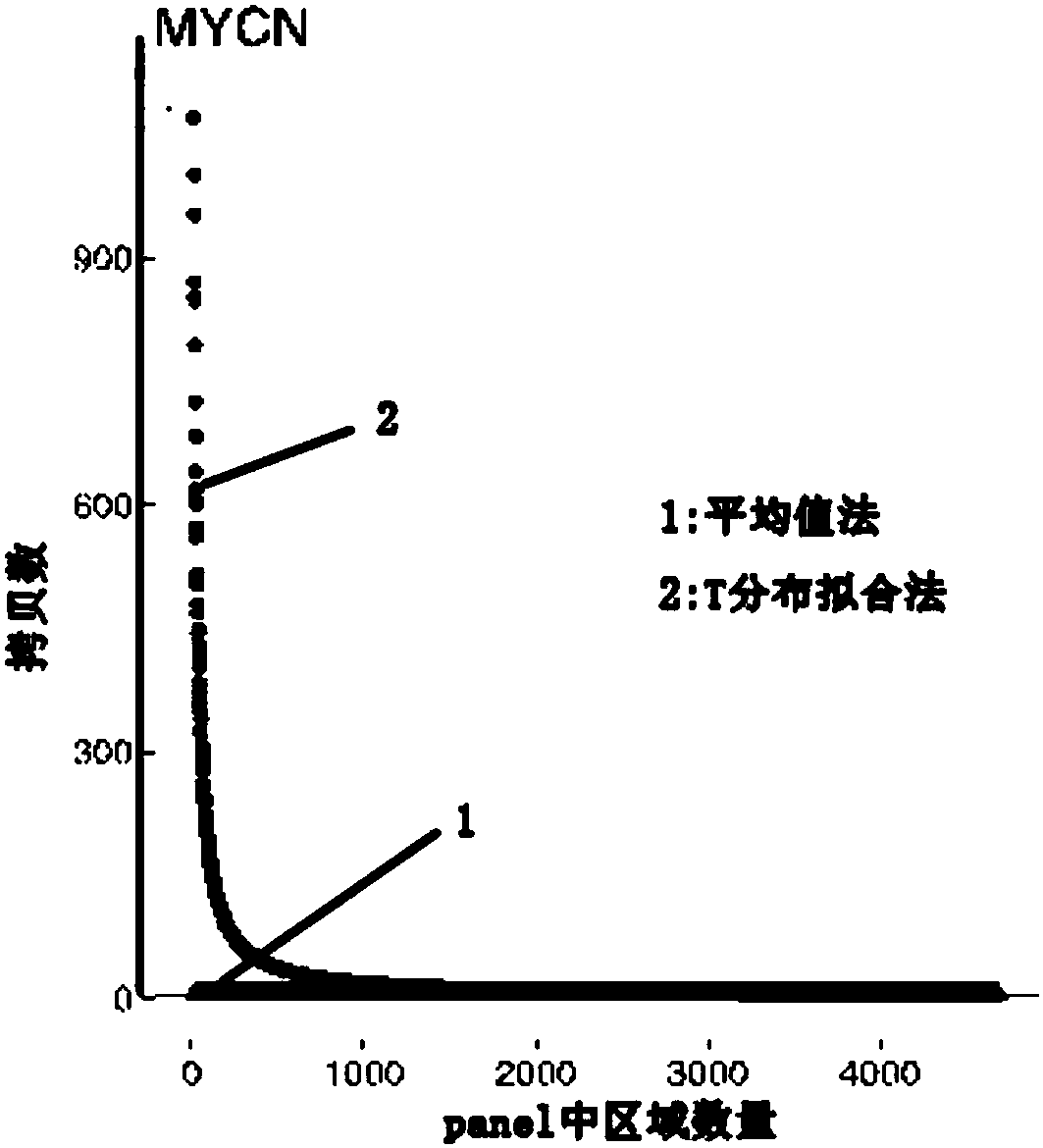
[0096] Example 1 Detection of MYCN gene copy number
[0097] The target region detected in this embodiment is to detect the copy number of the MYCN gene containing 2 exon regions, and the main information of the target region is shown in the following table:
[0098] Table 1 Two exons of MYCN gene
[0099]
[0100] There are a total of 349 normal people here. The mean value of the z-distribution curve composed of their MYCN gene OR values is 1.08, and the sd is 0.075. The patient samples here use the HD786 standard provided by https: / / www.horizondiscovery.com product, which contains MYCN amplification, the official copy number OR is 4.75, we use the NGS method and the algorithm described above to get the copy number OR of the gene is about 3.95. The control sample used here is the NA18535 standard cell.
[0101] The panel for detection also includes the exon regions of some other sensitive genes. Different numbers of regions are taken to form the panel together with the...
Embodiment 2
[0125] Example 2 Detection of MET gene copy number
[0126] The target region detected in this embodiment is to detect the copy number of the MET gene containing 20 exon regions, and the main information of the target region is shown in the following table:
[0127] table 5
[0128]
[0129] There are a total of 349 normal people here. The mean value of the z-distribution curve composed of the MET gene OR value is 1.01, and the sd is 0.039. The patient samples here use the HD786 standard provided by https: / / www.horizondiscovery.com product, which contains MET amplification, the official copy number OR is 2.25, we use the NGS method and the algorithm described above to get the OR of the gene is 1.72. The control sample used here is the NA18535 standard cell.
[0130] The panel for detection also includes the exon regions of some other sensitive genes. Different numbers of regions and 20 exons of the MET gene are selected to form a panel. A total of 4666 other candidate reg...
Embodiment 3
[0141] Example 3 Detection of CDKN2A gene copy number
[0142] The target region detected in this embodiment is to detect the copy number of the CDKN2A gene containing 5 exon regions, and the main information of the target region is shown in the following table:
[0143] Table 8
[0144]
[0145] There are a total of 349 normal people here. The mean value of the z-distribution curve composed of their CDKN2A gene OR value is 1.01, and the sd is 0.054. The patient samples here use the HD786 standard provided by https: / / www.horizondiscovery.com The official did not provide the copy number variation of this gene, but the OR of this gene obtained by using the NGS method and the algorithm described above is about 0.62.
[0146]The panel for detection also includes the exon regions of some other sensitive genes. Different numbers of regions are taken to form the panel together with the 5 exons of the CDKN2A gene. A total of 4681 other candidate regions are selected, for example, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com