A method for preparing long dna probes containing multiple repeating units
A technology of repeating units and long probes, applied in the field of oligonucleotide probe preparation, can solve the problems of limiting the magnification of bDNA structure and lower connection efficiency, and achieve the effect of increasing the magnification and increasing the sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
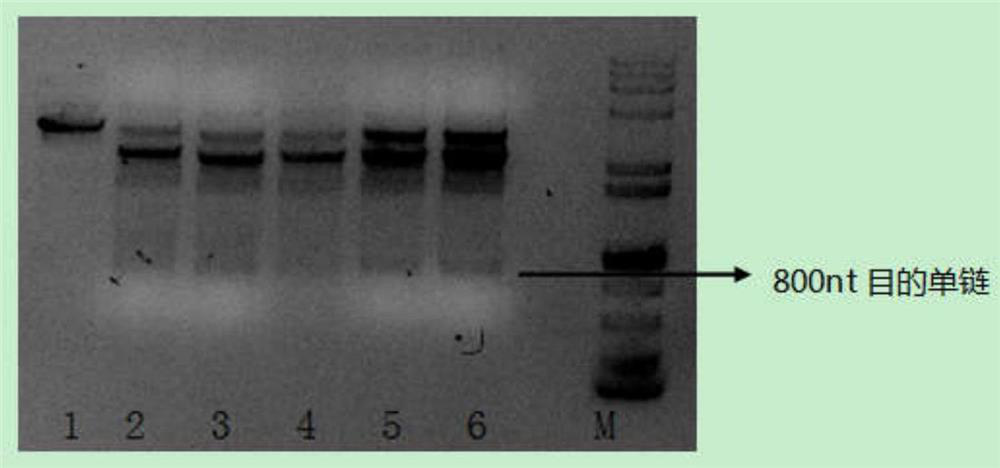
[0030] Preparation of 800nt, 20 repeat unit DNA long probes
[0031] (1) Preparation of a plasmid containing 20 repeating unit sequences, the specific steps are as follows:
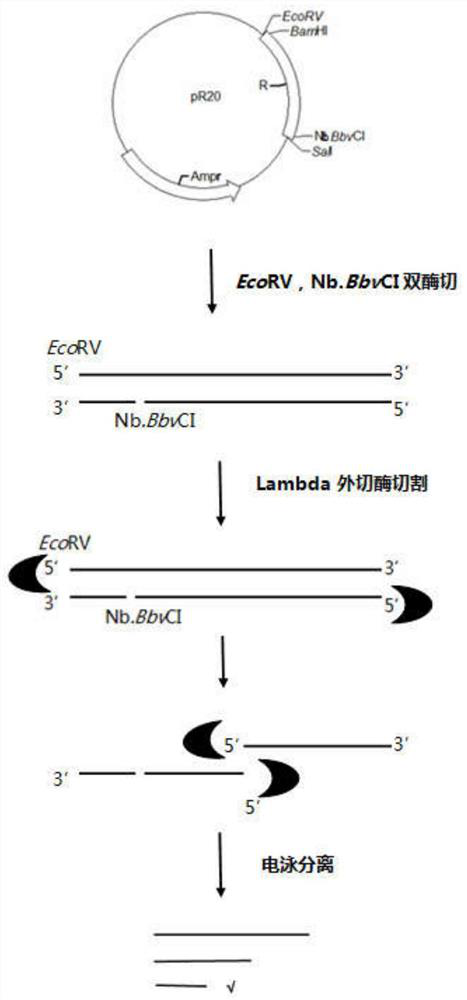
[0032] 1. Design 20 repeating unit sequences (sequence R, see Sequence Listing 1), with restriction sites EcoRV and Nb.BbvCI at both ends.
[0033] Synthesized by Sangon Bioengineering Co., Ltd. and linked into pUC57 to obtain a new plasmid pR20 containing the sequence R (attached figure 1 ) and stored in Escherichia coli.
[0034]2. Inoculate Escherichia coli containing pR20 in 3ml Amp LB medium, and culture at 37°C 200r / min for 16h.
[0035] 3. Use Axygen's plasmid mini-extraction kit (AP-MN-P-50) for plasmid extraction. (The steps are consistent with those provided in the kit)
[0036] 4. EcoRV (NEB R0195S), Nb.BbvCI (NEB R0631S) double-digest pR20, the specific system is:
[0037]
[0038] The reaction conditions are: 37°C, 2h; 80°C, 10min.
[0039] 5. Use Axygen's PCR Cleanup kit (AP-PCR-50G...
Embodiment 2
[0075] Preparation of 400nt, 10 repeat unit DNA long probes
[0076] (1) Preparation of a plasmid containing 10 repeating unit sequences, the specific steps are as follows:
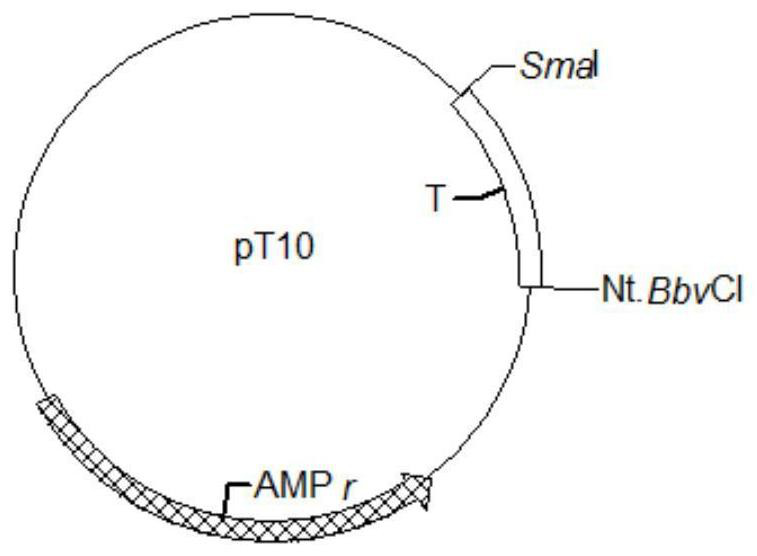
[0077] 1. Design 10 repeating unit sequences (sequence T, see Sequence Table 2), the restriction sites at both ends are SmaI and Nt.BbvCI respectively, synthesized by Sangon Bioengineering Co., Ltd. and ligated into pUC57. Obtain new plasmid pT10 containing sequence T (attached image 3 ) and stored in Escherichia coli.
[0078] 2. Inoculate Escherichia coli containing pT10 in 3ml Amp LB medium and culture at 37°C 200r / min for 16h.
[0079] 3. Use Axygen's plasmid mini-extraction kit (AP-MN-P-50) for plasmid extraction. (The steps are consistent with those provided in the kit)
[0080] 4. SmaI (NEB R0141V), Nt.BbvCI (NEB R0632S) double digestion of pT10, the specific system is:
[0081]
[0082] The reaction conditions are: 30°C, 2h; 80°C, 10min.
[0083] 5. Use Axygen's PCR Cleanup kit (AP-PCR-5...
Embodiment 3
[0105] Preparation of 200nt, 5 repeat unit DNA long probes
[0106] (1) Preparation of a plasmid containing 5 repeating unit sequences, the specific steps are as follows:
[0107]1. Design 5 repeating unit sequences (sequence F, see Sequence Table 3), the restriction sites at both ends are EcoRV and Nt.BbvCI respectively, synthesized by Sangon Bioengineering Co., Ltd. and ligated into pUC57. A new plasmid pF5 containing sequence F was obtained (attached Figure 4 ) and stored in Escherichia coli.
[0108] 2. Inoculate Escherichia coli containing pT10 in 3ml Amp LB medium and culture at 37°C 200r / min for 16h.
[0109] 3. Use Axygen's plasmid mini-extraction kit (AP-MN-P-50) for plasmid extraction. (The steps are consistent with those provided in the kit)
[0110] 4. EcoRV (NEB R0195S), Nt.BbvCI (NEB R0632S) double enzyme digestion pF5, the specific system is:
[0111]
[0112] The reaction conditions are: 37°C, 2h; 80°C, 10min.
[0113] 5. Use Axygen's PCR Cleanup kit ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com