Ensemble learning method for predicting DNA protein binding site
A combination site and integrated learning technology, applied in the field of bioinformatics, can solve the problem of not taking into account the data imbalance characteristics of DNA protein binding sites, so as to improve the effect of the model, improve the accuracy, and reduce the recognition error rate.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0022] In order to make the objectives, technical solutions and advantages of the present invention clearer, the present invention will be further described in detail below in conjunction with specific embodiments and with reference to the accompanying drawings. It should be understood that these descriptions are only exemplary and not intended to limit the scope of the present invention. In addition, in the following description, descriptions of well-known structures and technologies are omitted to avoid unnecessarily obscuring the concept of the present invention.
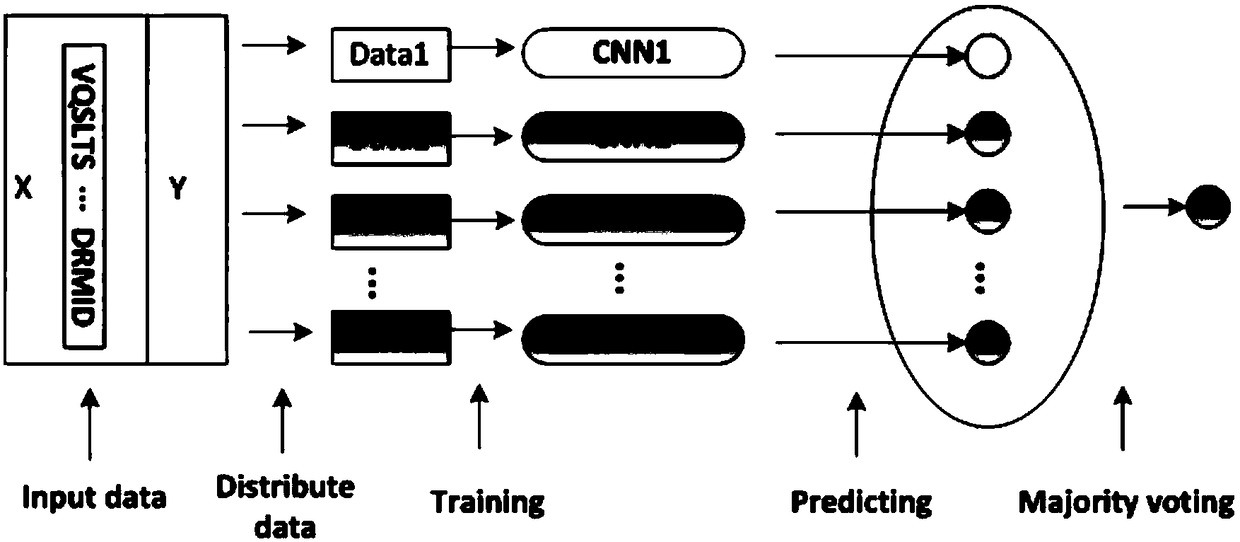
[0023] figure 1 Shows the schematic flow chart of the ENSEMBLE integrated learning method (ENSEMBLE-CNN) in the present invention. In the case of unbalanced data, the integrated learning method for predicting DNA protein binding sites of the present invention roughly includes the following steps:
[0024] S1) Obtain the protein sequence data of the DNA binding protein site;
[0025] S2) Preprocessing the protein seque...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com