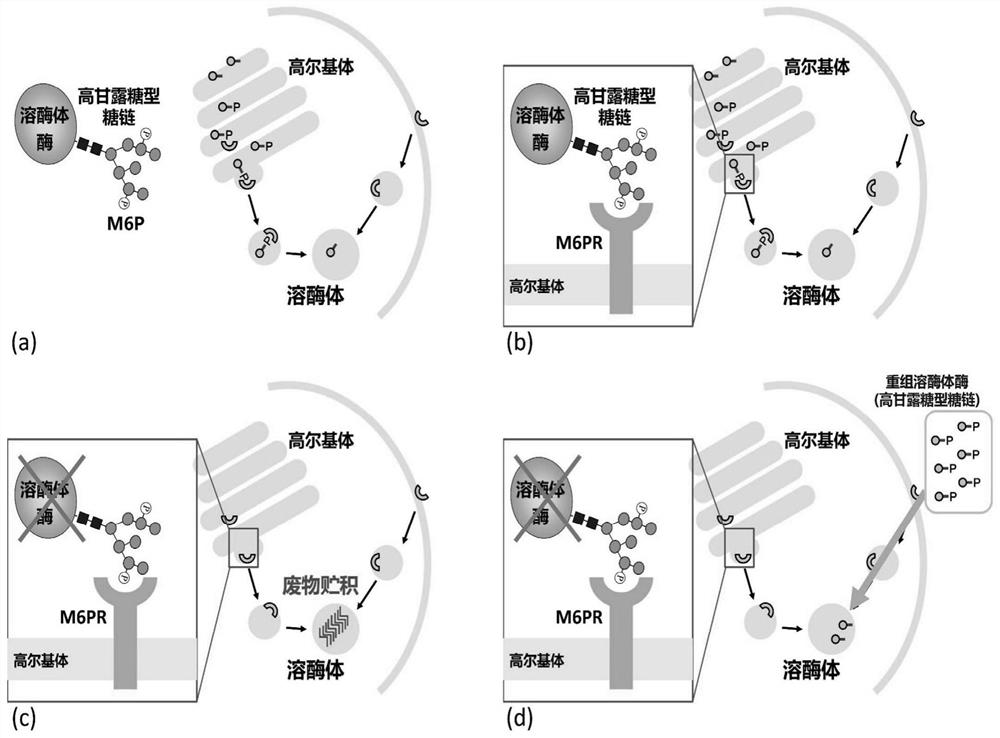
Animal cell line and method for producing glycoprotein, glycoprotein and use thereof
一种动物细胞、细胞株的技术,应用在生物化学设备和方法、遗传修饰的细胞、引入外来遗传物质而修饰的细胞等方向,能够解决糖链含量高、减弱效率、治疗效率不高等问题,达到含量降低、稳定性和安全性优异、糖链均一性高的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
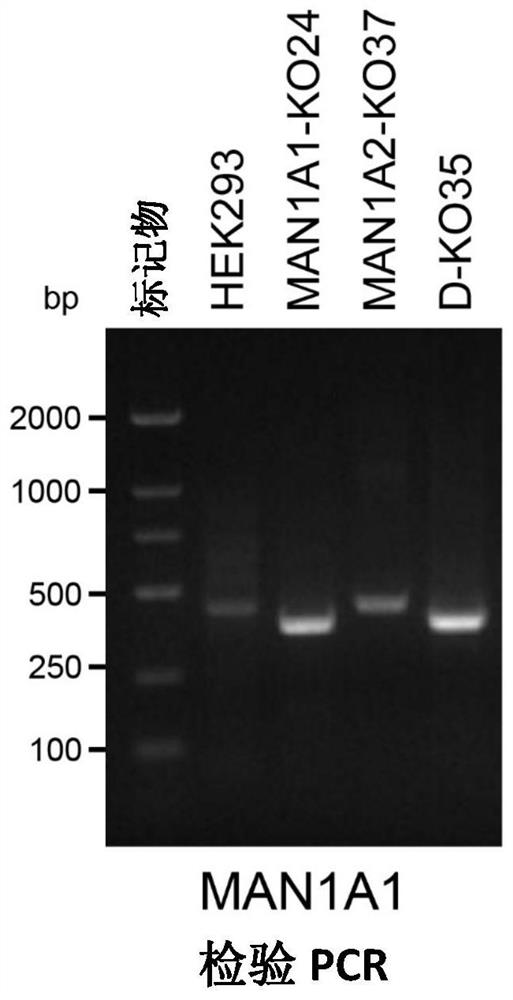
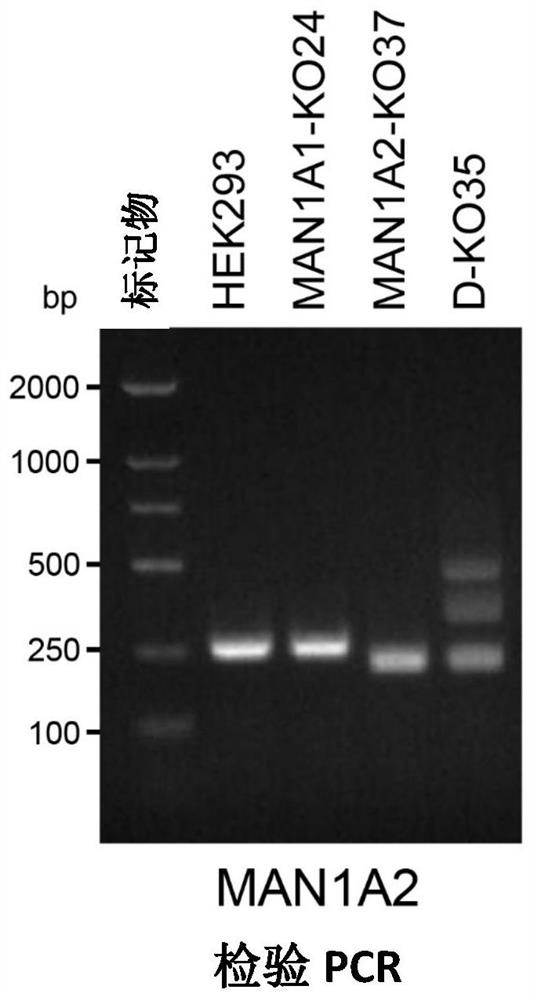
[0109] [Example 1] Using the CRISPR-Cas9 (Clustered Regularly Interspaced ShortPalindromic Repeats) system to construct a double knockout cell line of the Golgi α-mannosidase gene (mammalian cell line: human embryonic kidney cell HEK293).
[0110] 1. Construction of knockout plasmids
[0111] Gene knockout using CRISPR-Cas9 technology often requires the design of a 20bp sequence fragment, and there must be a PAM site (NGG / NAG) behind the sequence fragment. In this experiment, the gene sequences of the two genes MAN1A1 / MAN1A2 to be knocked out were downloaded from NCBI (see SEQ ID NO: 44 and SEQ ID NO: 45, respectively). About the design of guide-RNA, in Michael Boutros lab's Target Finder ( http: / / www.e-crisp.org / E-CRISP / designcrispr.html ) to find the DNA sequence of the guide-RNA required to knock out the gene.
[0112] The two target sequences of MAN1A1 and the primer sequences used for each are:
[0113] MAN1A1KO1: AAAACCACGAGCGGGCTCTCAGG (serial number 1)
[0114]...
Embodiment 2
[0147] [Example 2] Analysis of sugar chains on the cell surface using a flow cytometer
[0148] After using the CRISPR / Cas9 system to knock out MAN1A1 / MAN1A2, two synthetic genes that control α-mannosidase in the Golgi apparatus, the sugar chains on the surface of the double knockout cell line will change to a certain extent. We confirmed this phenomenon with two different fluorescently labeled lectins. The lectin PHA-L4-FITC can recognize complex sugar chains on the cell surface, and the lectin ConA-FITC can recognize high-mannose sugar chains on the cell surface. The type of sugar chains on the surface of different cell lines can be compared by staining the cells with lectin. The specific method is as follows:
[0149] (1) Inoculate different cell lines in 6-well plates and wait for their growth to be 100%
[0150] (2) Remove the medium and wash once with 1ml of PBS
[0151] (3) Add 220ul Tryp / EDTA to digest the cells
[0152] (4) Add 1ml of fresh 10% FCS medium to harve...
Embodiment 3
[0166] [Example 3] Knockout of other genes related to α1,2-mannosidase
[0167] Introduce knockout plasmids of the two genes MAN1C1 and MAN1B1 into DKO cells (for the two knockout target sequences of the MAN1C1 gene and the corresponding primer sequences, see SEQ ID NO: 13-18, the two knockout target sequences of the MAN1B1 gene and the corresponding See sequence numbers 19-24 for the primer sequences respectively), and the plasmids introduced into the cells will express the sequences of Cas9 protein and target RNA. Cells were cultured for about ten days after transfection, and the cell genome was extracted. Because two target sequence sites are designed, the gene sequence on the chromosome will be shifted after knocking out the gene, and the inventors have confirmed the knockout of some genes (results shown in Figure 8, sequence numbers 29 and 30 are used for PCR testing of MAN1C1 Gene primers, sequence numbers 31 and 32 represent primers for PCR detection of MAN1B1 gene), a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com