Separation of satellite DNA pBv sequence of centromere of halogeton glomeratus and application thereof
A halophyte and centromere technology, applied in the field of plant science, can solve the problems of lack of and limited species research depth and utilization value, and achieve highly targeted and specific effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] This embodiment provides a method for isolating the centromere satellite DNA pBv sequence of Grass saline chromosome, which specifically includes the following steps:
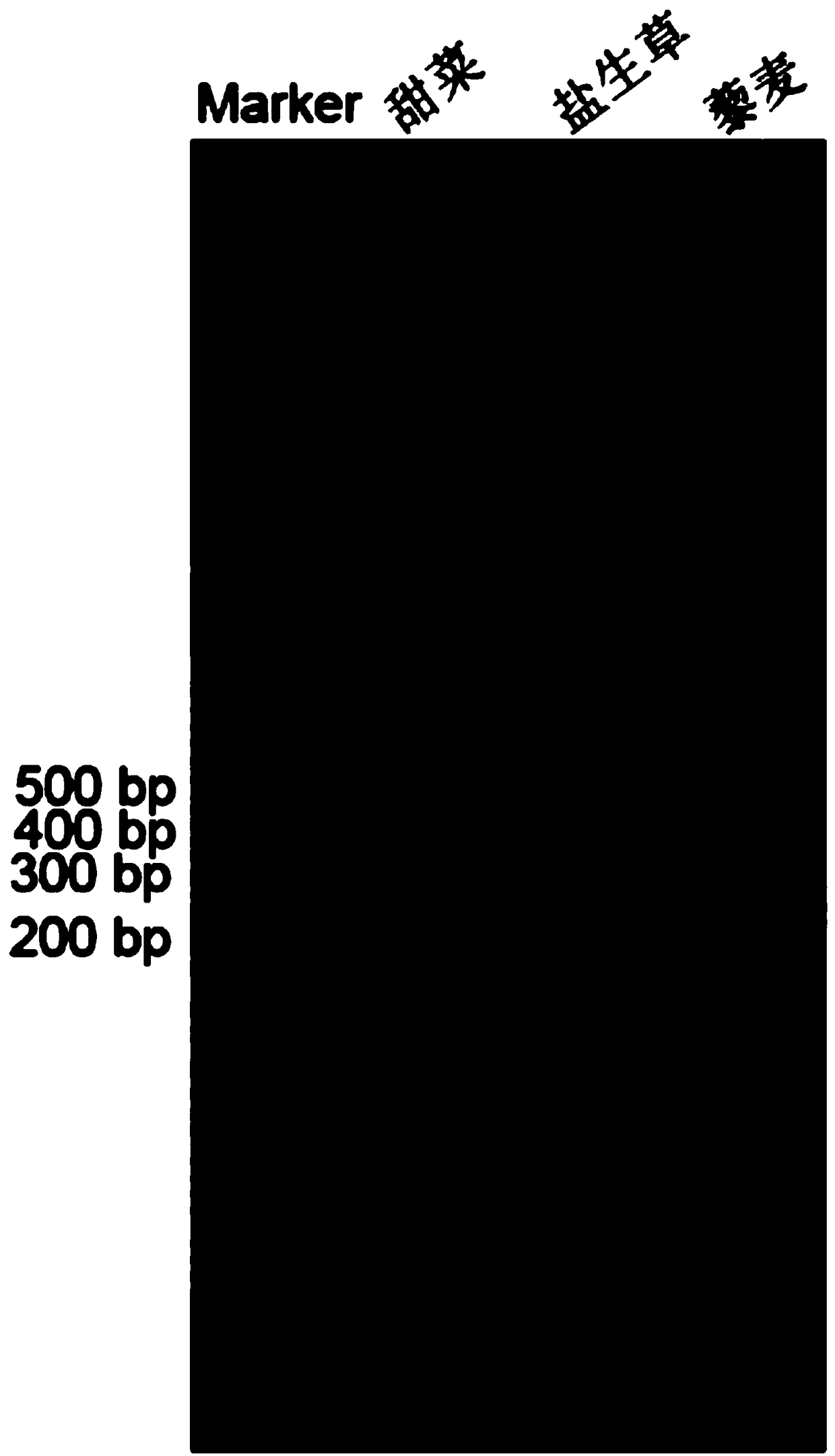
[0030] Primers were designed according to the conserved motifs of the chromosome centromere satellite DNA pBv sequence in plants, and the genomic DNA of Halophylla saline was extracted, and the genomic DNA was used as a template for PCR amplification to obtain the target fragment ( figure 1 ), and then recover the target fragment, and connect it to the pMD19-T cloning vector, transform Escherichia coli DH-5a competent cells, screen the positive clones, and extract the plasmid of the appropriate bacterial liquid through PCR bacterial liquid detection and send it to BGI for Sequencing analysis obtained the centromere satellite DNA pBv sequence of Grass saline, specifically as shown in SEQ ID No: 1, with a size of 327 bp ( figure 1 ):
[0031] The primer sequences used in this step are: upstream primer F...
Embodiment 2
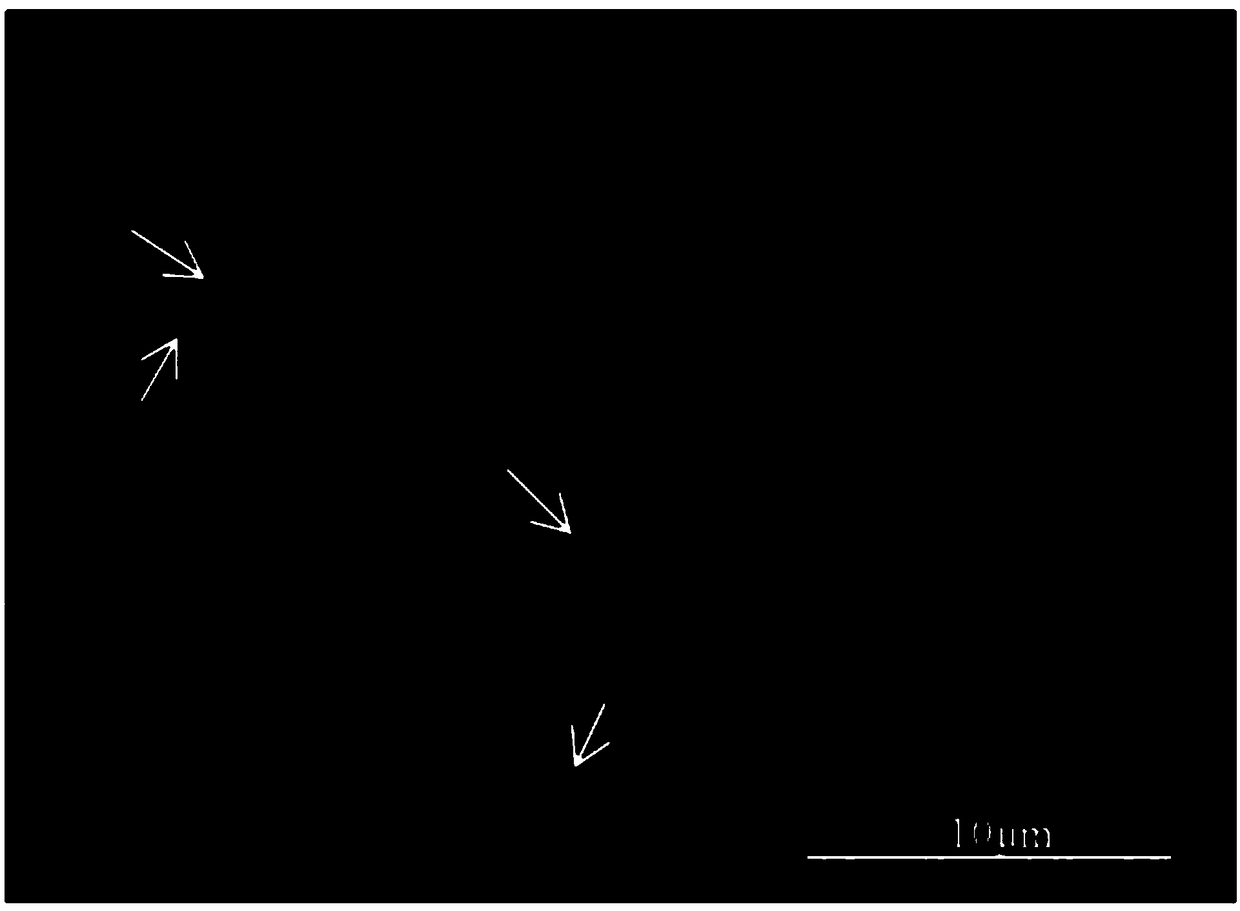
[0034] This embodiment provides the application of the centromere satellite DNA pBv sequence of the Chromosome DNA pBv as a molecular marker in the analysis of species genetic variation and evolution analysis, specifically including the following steps:
[0035] (1) Preparation of digoxigenin-labeled probes
[0036] According to Example 1, the plasmid DNA containing the centromere satellite DNA pBv sequence element of Grass saline was obtained as the probe DNA, and the DIG-Nick Translation Mix Digoxigenin Labeling Kit was used for labeling. The reaction system was 1 μL of the probe DNA, Nick Translation Mix 4 μL. Mark in a constant temperature water bath at 15°C for 1.5 h-2.0 h, then freeze at -20°C in the dark.
[0037] (2) Preparation of metaphase chromosomes in Grass saline
[0038] Place the halophytic grass seeds treated with swollenin in a petri dish and cultivate them in the dark at 28°C. When the roots grow to about 1-1.5cm, remove the root tips and treat them in a m...
Embodiment 3
[0042] This embodiment provides the application of the centromere satellite DNA pBv sequence in the species identification of Grass saline chromosome, which specifically includes the following steps:
[0043] (1) Cloning and sequencing of the pBv sequence of Grass salina
[0044] Primers were designed according to the conserved motifs of the chromosome centromere satellite DNA pBv sequence in plants, and the genomic DNA of Halophylla saline was extracted, and the genomic DNA was used as a template for PCR amplification to obtain the target fragment ( figure 1 ), and then recover the target fragment, and connect it to the pMD19-T cloning vector, transform Escherichia coli DH-5a competent cells, screen the positive clones, and extract the plasmid of the appropriate bacterial liquid through PCR bacterial liquid detection and send it to BGI for Sequencing analysis obtained the centromere satellite DNA pBv sequence of Grass saline, specifically as shown in SEQ ID No: 1, with a siz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com