Sequencing data mutation analysis system
A technology for analyzing systems and sequencing data, applied in the field of biomedicine, which can solve problems such as low number of variants and false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0157] Example 1 A sequencing data mutation analysis system
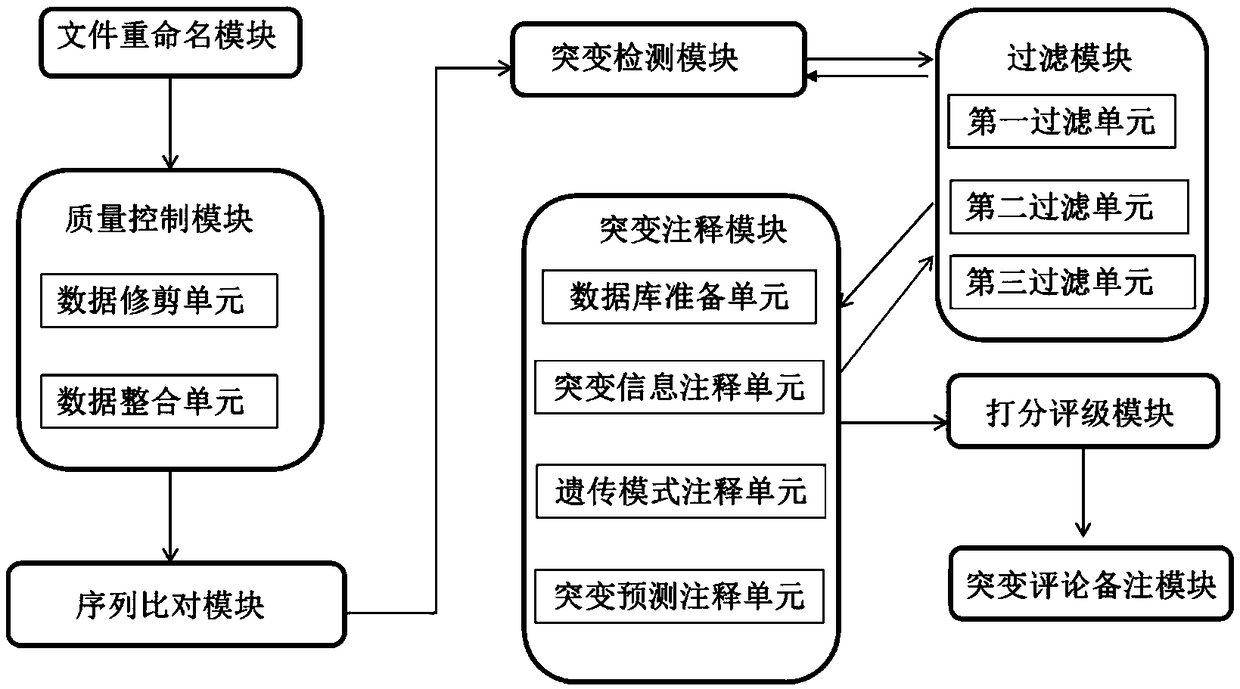
[0158] A sequencing data mutation analysis system, the analysis system includes a file renaming module, a quality control module, a sequence comparison module, a mutation detection module, a mutation annotation module, a scoring and rating module, a filtering module, and a mutation comment comment module;
[0159]The file renaming module is used to unify the sequence number into the analysis number, merge multiple fastq files of the same sample to establish a corresponding table of sequence number and sample number, and name it id.dic. Through the Python script, the fastq file named after the sequence number The file is renamed to a fastq file named after the sample number (or other analysis numbers). At the same time, if a sample is used on the machine multiple times or on multiple lanes, it can be automatically merged, as long as the sample numbers are consistent.
[0160] The quality control module data pruning u...
Embodiment 2
[0219] Embodiment 2 specific running example
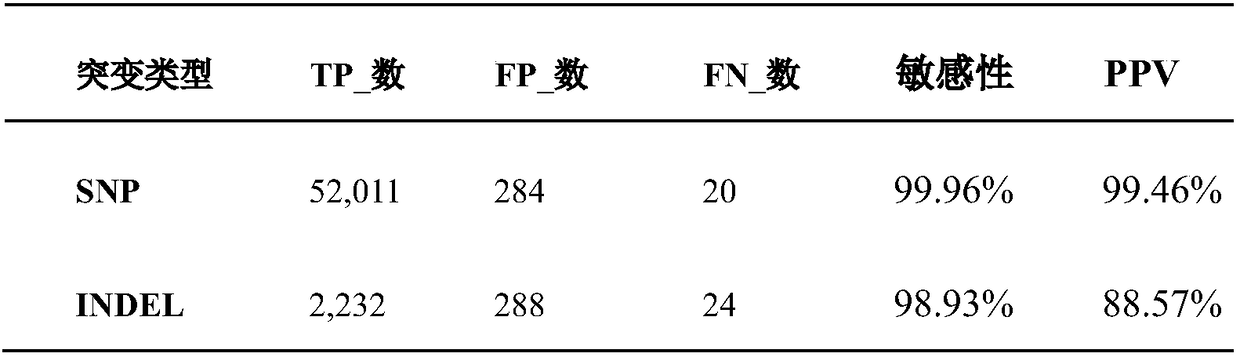
[0220] 1. Data introduction
[0221] Data Type: Whole Exome Sequencing
[0222] Tissue source: DNA from blood of brain arteriovenous malformation (BAVM) patients and their parents
[0223] Experimental Design: Exon Capture Sequencing
[0224] Sequencing platform: Illumina HiSeq 4000
[0225] 2. System use
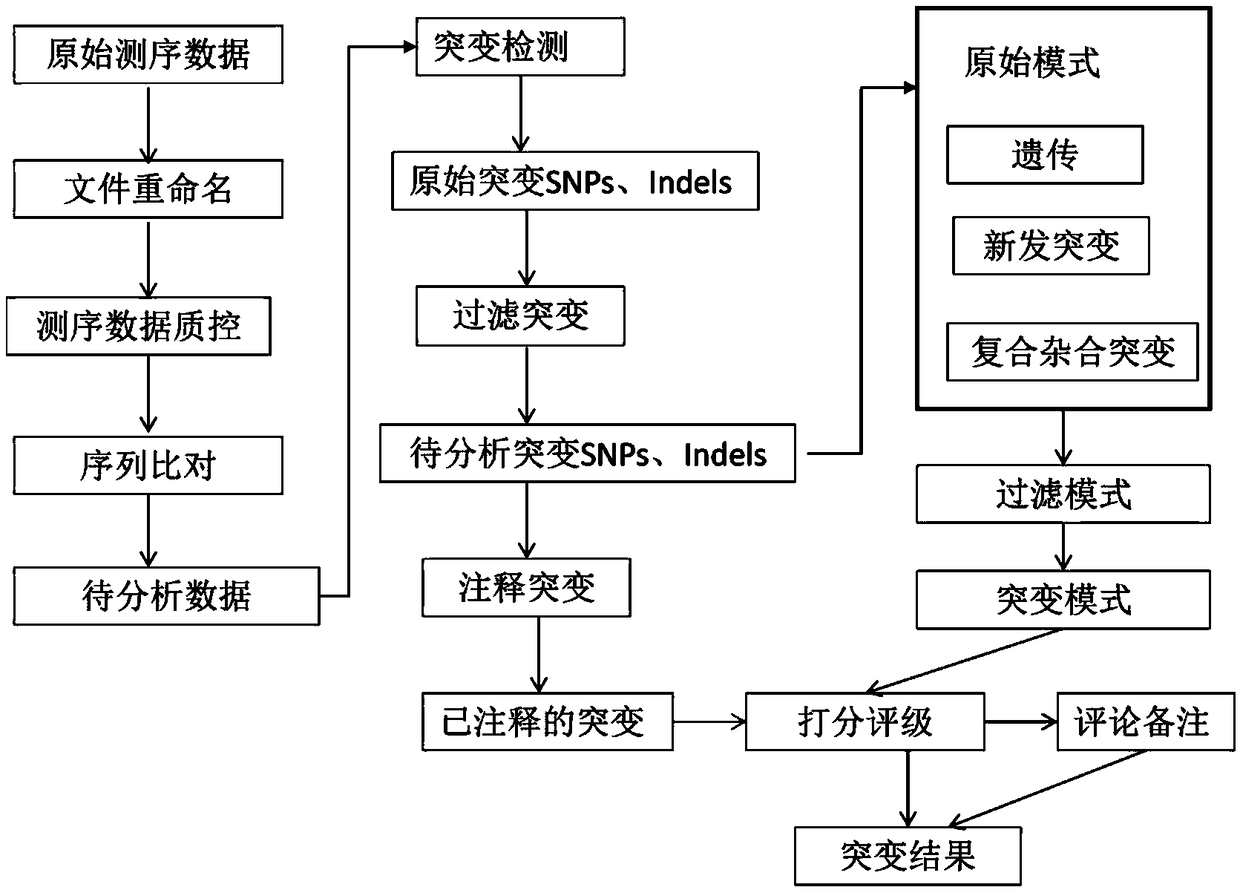
[0226] The whole exome velocimetry data analysis process is as follows: figure 2 The process shown includes: renaming of sequencing data, quality assessment and control of sequencing data, detection and annotation of mutations, scoring and comments of mutations, etc. Next, use the function modules integrated in the software to realize each analysis step step by step:
[0227] 1) Use the file renaming module to name the sequencing number as the analysis number, unify the naming format, and merge multiple fastq files of the same sample to establish a correspondence table between the sequencing number and the sample number;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com