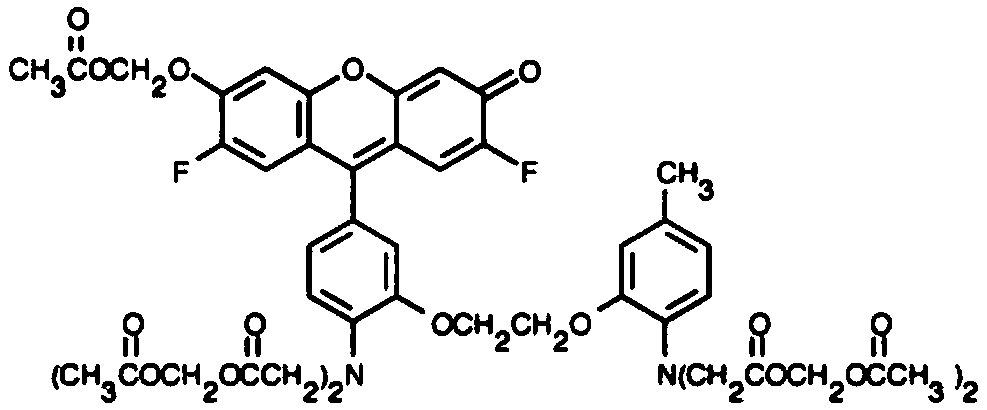
Quantitative method for determining sweetness at cellular level
A measurement method and quantitative measurement technology, applied in the field of molecular biology, can solve the problems of strong subjectivity in manual evaluation, inability to quantify sweetness, and inability to accurately quantify sweetness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] Changes of intracellular calcium ion concentration in HEK293 cells and HEK293-T1R2 / T1R3 cells under 150mM sucrose solution:
[0052] (1) Planting cells in a confocal culture dish:
[0053] Incubate the special confocal culture dish (outer diameter 35mm, inner diameter 10mm) with 1ml polylysine (25μg / ml) at 37°C for 30min, wash with phosphate buffer (10mM, pH 7.4) three times, and passage the cells in proportion into the confocal dish. After 24h-48h of adherent growth of the cells in the culture dish, it is observed under an optical microscope that the area of the cells covering the bottom of the culture dish reaches 40%-60%.
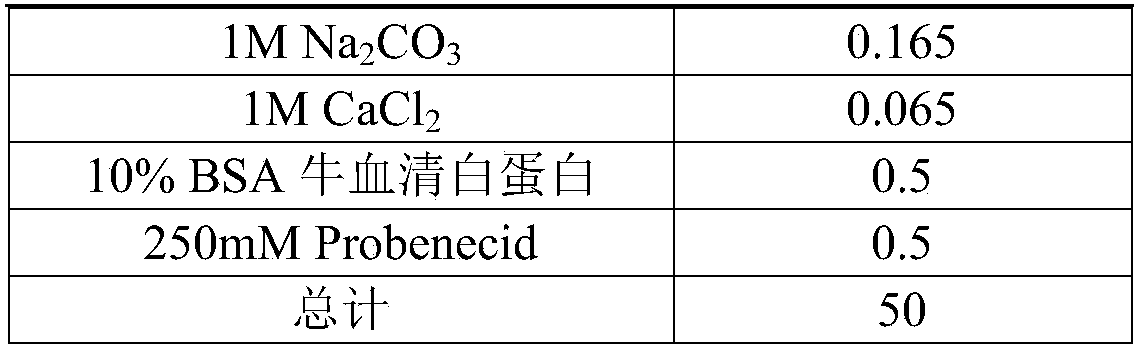
[0054] (2) Configure Flex buffer (pH=7.4)
[0055] Reagent
Volume (mL)
HBSS solution
47.5
HEPES solution
1
1M MgSO 4
0.05
1M Na 2 CO 3
0.165
1M CaCl 2
0.065
10% BSA bovine serum albumin
0.5
250mM Probenecid
0.5
total
50
[0056] The HBSS ...
Embodiment 2
[0076]Under the stimulation of 50 mM sucrose, the intracellular calcium ion concentration of HEK293 cells and HEK293-T1R2 / T1R3 cells changed, and the steps and conditions were the same as those in Example 1.
[0077] The fluorescence change ratio ΔR was obtained to be 0.105. If the sweetness of the 150 mM sucrose solution in Example 1 is set to 1, then the ΔR / ΔRs value is 0.370, and the relative sweetness of the 50 mM sucrose solution in this embodiment is 0.370.
Embodiment 3
[0079] Under the stimulation of 250 mM sucrose, the intracellular calcium ion concentration of HEK293 cells and HEK293-T1R2 / T1R3 cells changed, and the steps and conditions were the same as those in Example 1.
[0080] The fluorescence change ratio ΔR was obtained to be 0.487. Assuming that the sweetness of the 150mM sucrose solution in Example 1 is 1, the ΔR / ΔRs value is 1.71, and the relative sweetness of the 250mM sucrose solution in this example is 1.71.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com