SiRNA interfering with HSPA13 expression and application thereof
A gene expression and molecular technology, applied to siRNA that interferes with HSPA13 expression and its application field, can solve the problem of not studying the EMT process of HSPA13 epithelial cells, and achieve the effect of reducing recurrence and avoiding risks.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
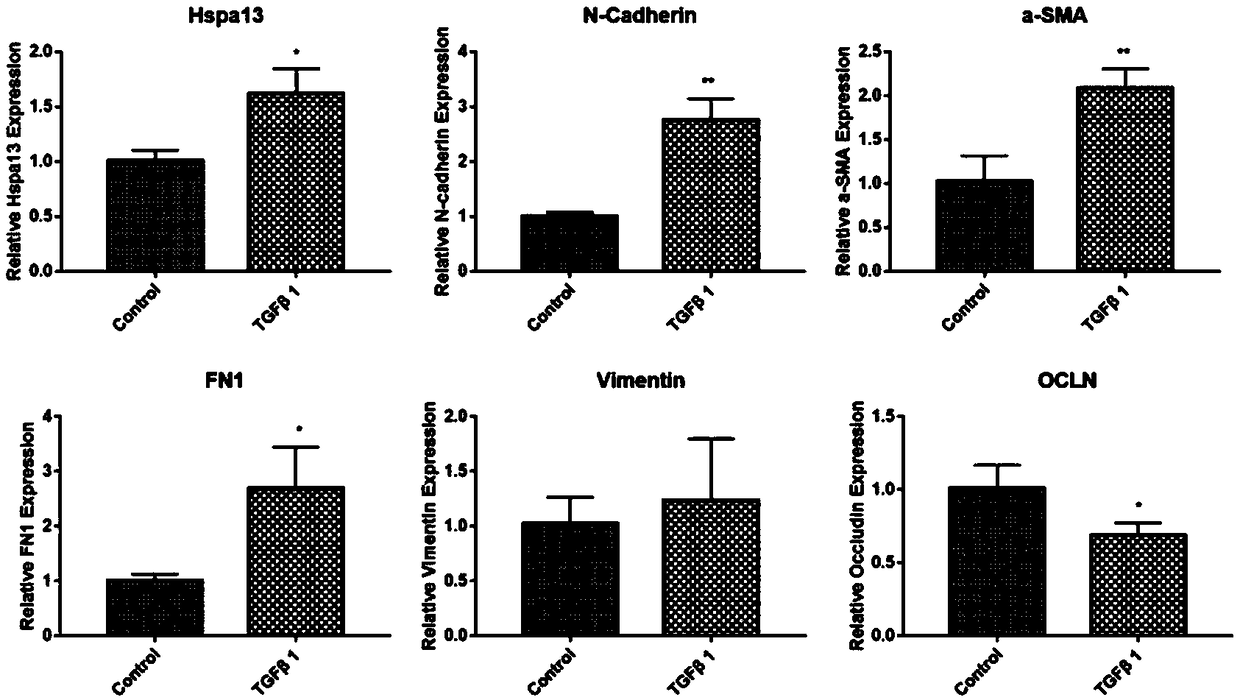
[0048] Establishment of EMT cell model in vitro to detect the expression level of HSPA13
[0049] Establish an in vitro EMT cell model: when the confluence of ARPE19 cells is about 60-70%, treat with serum-free medium overnight, and then treat ARPE19 cells with 10ng / ml TGFβ1 to induce EMT in ARPE19 cells. After induction, the morphology of RPE cells changed from epithelial to fibroblastic, while loss of tight junctions and accelerated cell migration.
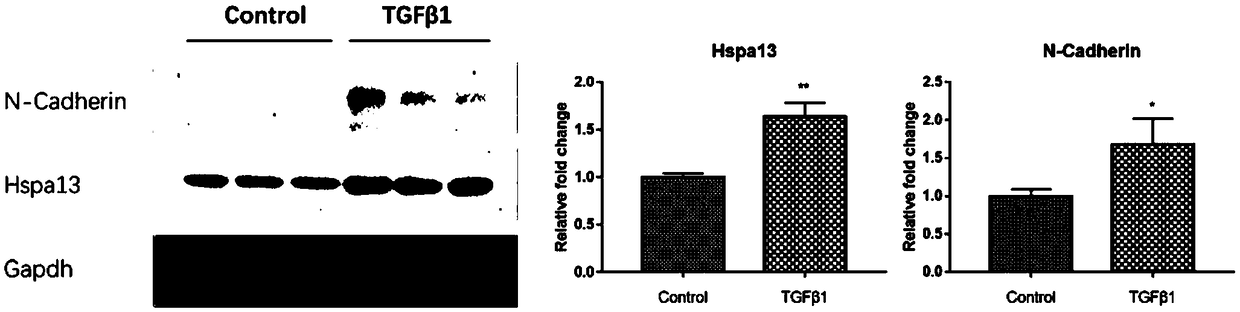
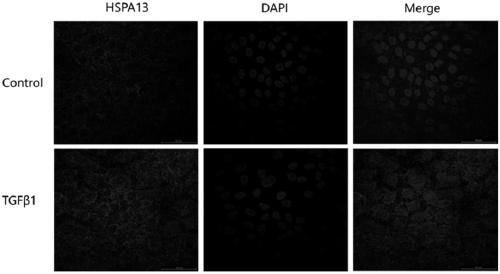
[0050] Q-PCR, Western blot and immunofluorescence to detect the expression level of HSPA13
[0051] Q-PCR, Western blot and immunofluorescence were used to detect the expression level of HSPA13 in ARPE19 cells before and after induction.
[0052] (1) Q-PCR detection
[0053] The normal control and TGFβ1-induced cells were collected in a 1.5ml EP tube with 1ml Trizol, and the total RNA was extracted. After reverse transcription and real-time quantitative PCR analysis, it was calculated by semi-quantitative method.
[0054] Th...
Embodiment 2
[0095] Expression of HSPA13 and EMT-related genes detected after plasmid transfection and TGFβ1 induction
[0096] 1. Cell preparation: Inoculate the cells in a 6cm Petri dish, and when the confluence of the cells reaches 60-70%, starve the cells overnight with serum-free medium.
[0097] 2. The siRNA molecule has the sequence shown in SEQ ID NO.17, namely GGCUGACGUCUUCCACGUC. The long hairpin DNA molecule corresponding to the siRNA molecule was annealed in vitro and then ligated with the empty plasmid pSuper digested with BglII and Hind III, namely the plasmid HSPA13siRNA, and its sequence was verified by small-scale extraction and sequencing of the plasmid.
[0098] 3. Plasmid transfection: Invitrogen's Lipofectamine 3000 transfection reagent was used for plasmid transfection. The prepared empty plasmid pSuper and plasmid HSPA13siRNA were operated according to the instructions of the transfection reagent.
[0099] 4. Treat with TGFβ1 48h after transfection.
[0100] 5. Co...
Embodiment 3
[0104] Lentiviral infection and flow screening
[0105] 1. Cell treatment: seed the cells in a 6-well plate, and perform corresponding cell treatment (as described in step 1 of Example 2);
[0106] 2. Infection: Use the appropriate concentration and volume of lentivirus, add it to the serum-free cell culture medium, make a mark, and replace it with a complete medium containing 10% fetal bovine serum four hours later.
[0107] 3. Microscopic observation: 36 hours after infection, observe the green fluorescence under a fluorescent microscope, that is, whether there is expression of green fluorescent protein.
[0108] 4. Flow cytometric screening: After the green fluorescence is observed, perform flow cytometric screening. The specific steps are roughly described as follows: After trypsinizing and centrifuging the cells to be screened, use a 0.7um filter to disperse the cells into individual cells and collect them in the flow cytometry. Place the cells in a BD tube and put them ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com