Genetic marker for risk assessment on AML (acute myelogenous leukemia) and application of genetic marker
A technology for acute myeloid and leukemia, applied in the field of diagnosis, which can solve the problem that predictors have no prognostic effect, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
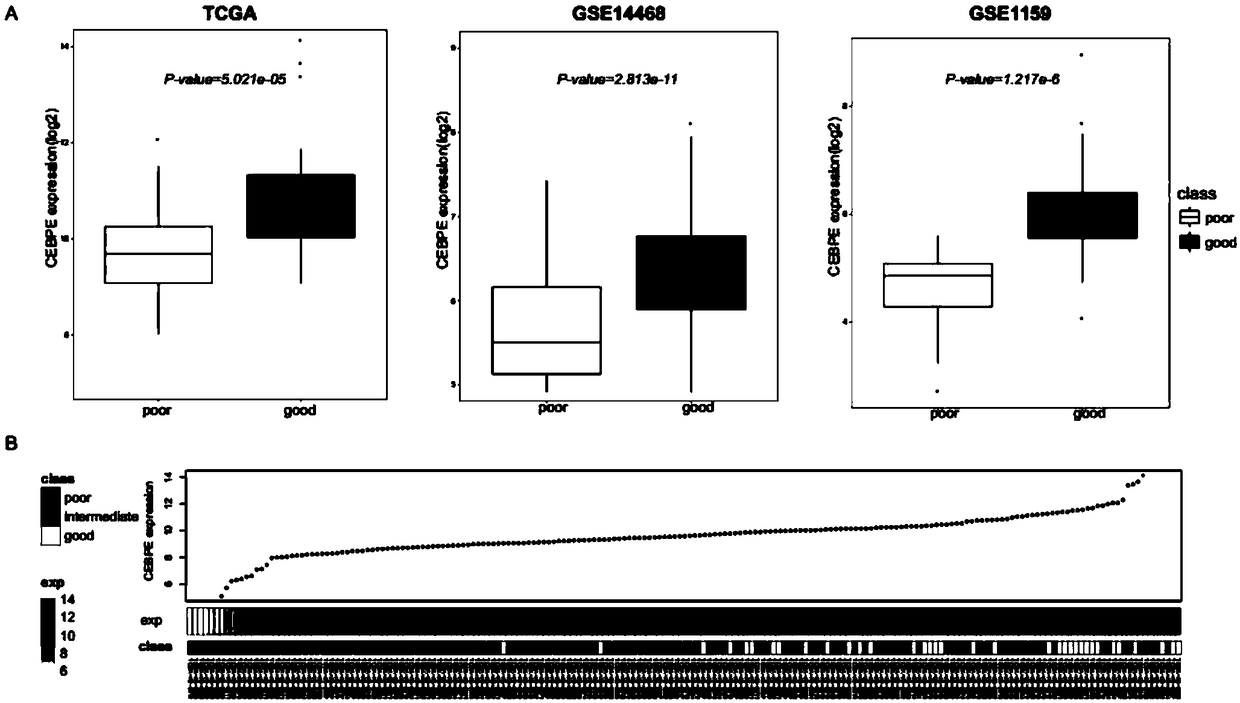
[0206] Example 1, CEBPe expression is significantly high in known AML patients with better prognosis
[0207] The present inventors collected AML patient expression profile data including prognostic classification information from public databases. Among them, there are 184 samples in TCGA, 186 samples in GSE14468, and 260 samples in GSE1159. The inventors found that in these three independent data sets, CEBPe was significantly overexpressed in samples known to have a better prognosis. The P values of differential expression of CEBPe in samples with good prognosis and poor prognosis were 5.021e-05, 2.813e-11, and 1.217e-6, respectively (see figure 1 A).
[0208] In the TCGA data, most of the samples with the highest expression of CEBPe had a better prognosis (see figure 1 B).
Embodiment 2
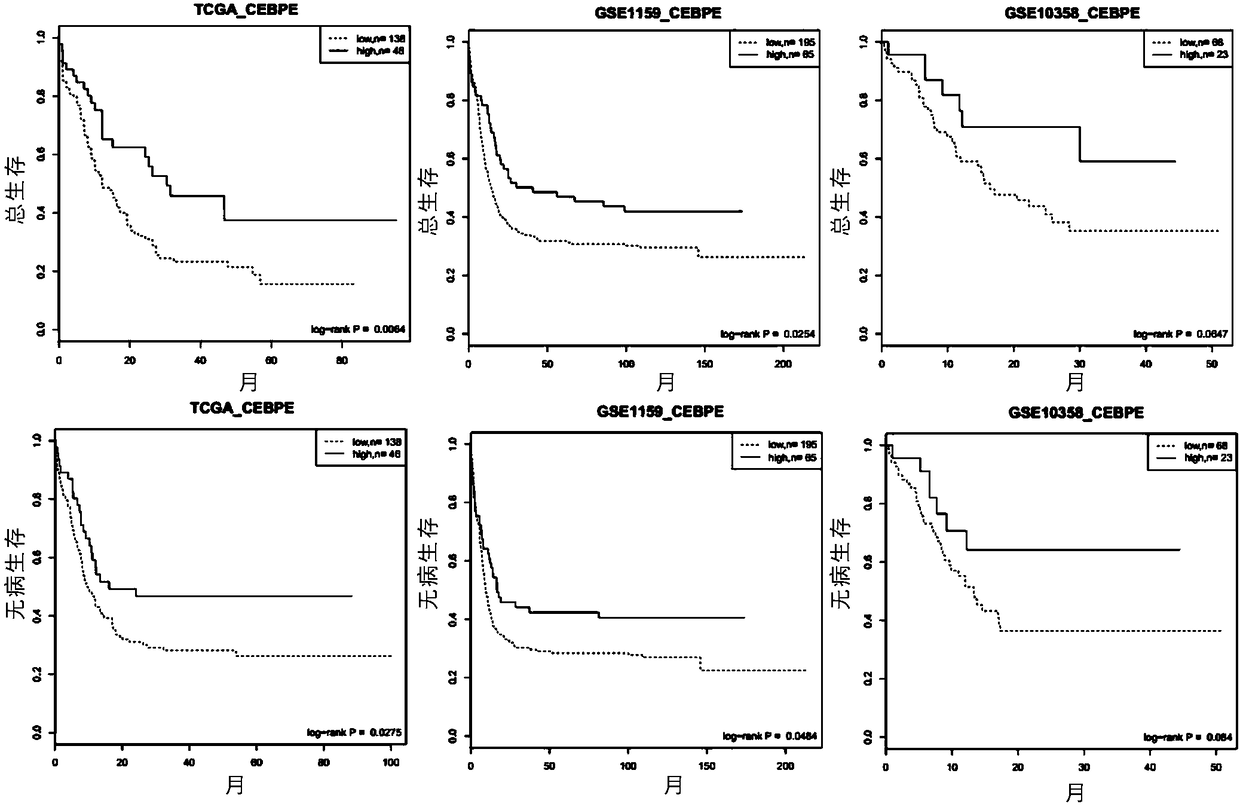
[0209] Example 2, CEBPe expression has a predictive effect on overall survival and disease-free survival
[0210] In three independent data sets of TCGA, GSE1159, and GSE10358, all samples in each data set were sorted according to the expression value of CEBPe from high to low. The samples with the top quartile of CEBPe expression values were classified as CEBPe high-expressing samples, and the remaining three-quarters of samples were classified as CEBPe-low expressing samples.
[0211] The results showed that the survival time of CEBPe high expression group was significantly longer than other samples. In TCGA, GSE1159, and GSE10358, the overall survival rates of high / low expression samples were 38% / 17%, 41% / 23%, 59% / 35%; the disease-free survival rates were 47% / 23%, 40% % / 21%, 62% / 37%. CEBPe's ability to predict overall survival and disease-free survival in three sets of independent data sets was significant (see image 3 ).
Embodiment 3
[0212] Example 3, CEBPe expression can predict the recurrence of AML
[0213] The present inventors used two sets of data sets, TCGA and GSE1159, which contain patient recurrence information, to evaluate the predictive ability of CEBPe expression for recurrence in AML patients. By using the K-nearest neighbor classification method, all samples were divided into CEBPe high expression group and low expression group, and then the recurrence of samples in the two groups was compared.
[0214] The results showed that CEBPe expression had a significant predictive effect on the recurrence rate of patients (see Figure 4 ). The recurrence rate of samples with high expression of CEBPe was significantly lower than that of samples with low expression.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com