Method for enhancing acid resistance of L-asparaginase
A technology for asparaginase and construction method, which is applied in the field of enhancing the acid resistance of L-asparaginase, and can solve problems such as inactivation and protein variation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
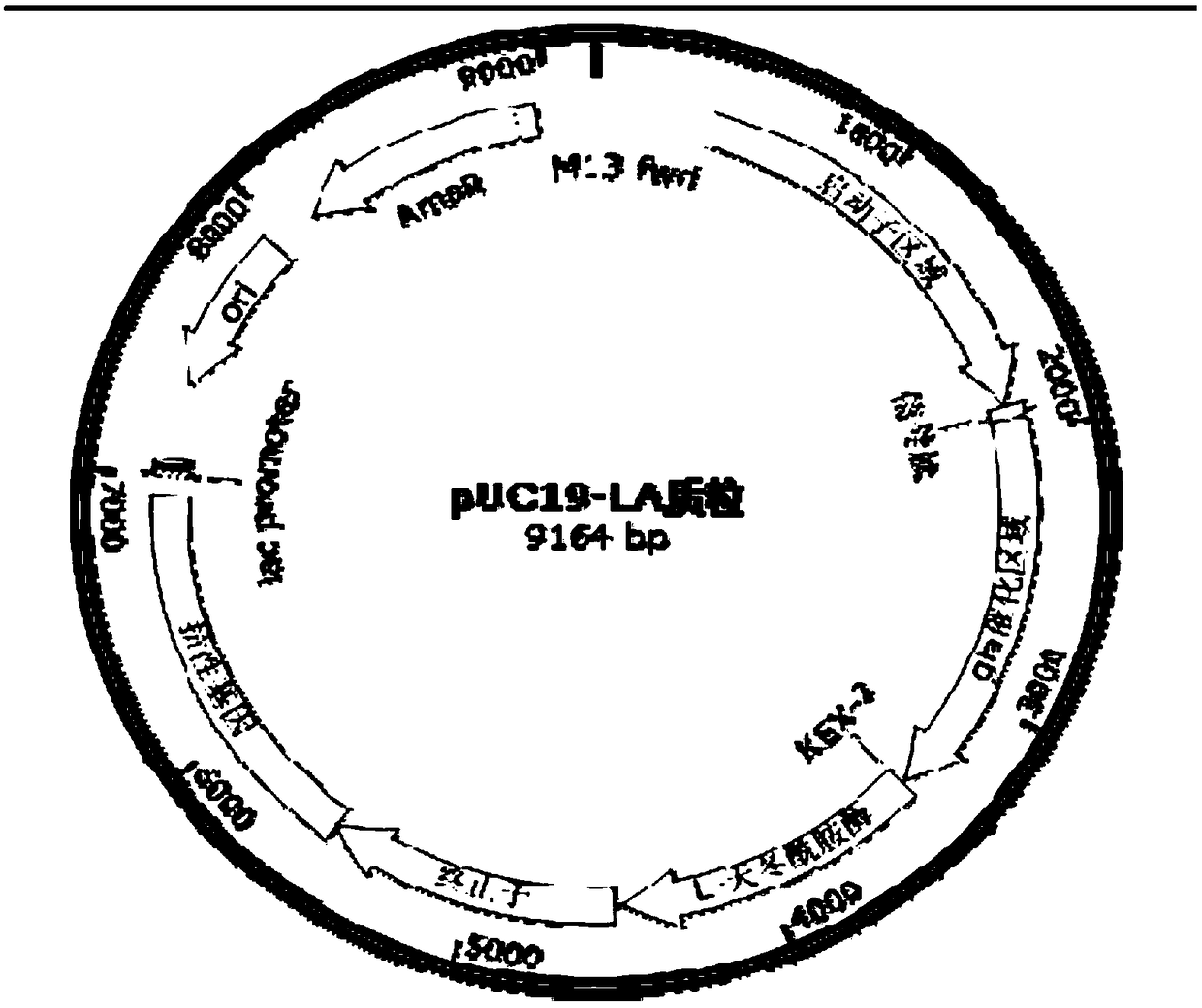
[0036] Example 1 Construction of recombinant plasmid fusion expressing L-asparaginase
[0037] Select the Aspergillus promoter Pgla, PgpdA or PaclA, and add 20bp upstream and downstream homology arms of the original promoter to the 5'and 3'ends of the selected promoter sequence (the upper and lower homology arm sequences are as SEQ ID NO. 5 and SEQ ID respectively). Shown in NO.6).
[0038] The markers include hygromycin B (hyg), orotidine-5'-phosphate dehydroxylase, acetamidase and other filamentous fungal markers with similar functions commonly used in Aspergillus. The hygromycin resistance gene in the recombinant plasmid is derived from the PAN7-1 plasmid, and the expression box primers are as follows (Hyg-F / R, see Table 1). If you want to choose other resistances, you can replace hyg in the expression box for construction.
[0039] Table 1 Primer list
[0040] Primer name
Primer sequence
Hyg-F
GAATTCCCTTGTATCTCTACACACAG
Hyg-R
TGAAGAACGAATACCGCGACATCCAACCCATC
[0041] ...
Embodiment 2
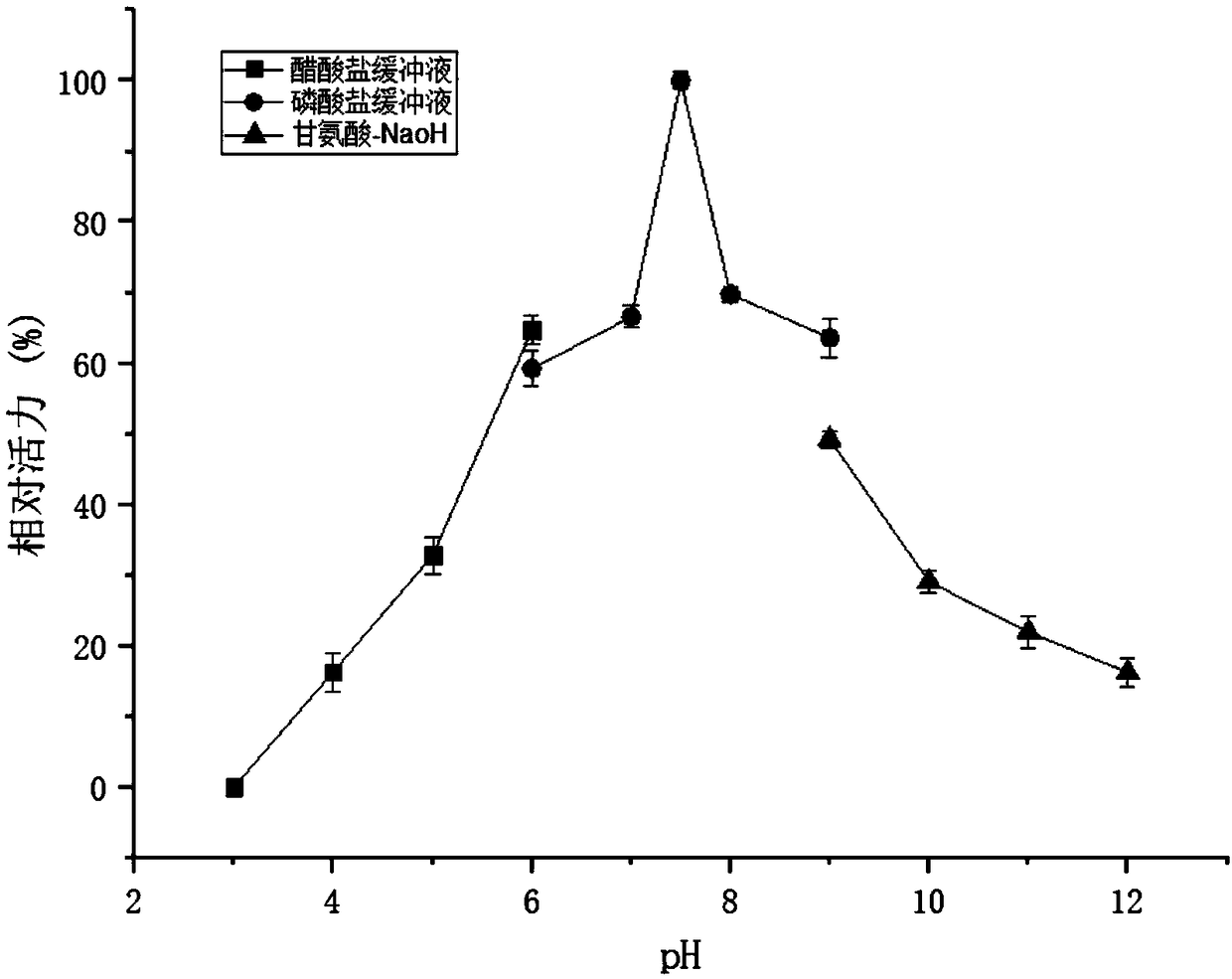
[0045] Example 2 Changes in enzyme properties of recombinant L-asparaginase
[0046] The obtained recombinant Aspergillus niger was inserted into YPM medium, and fermented for 72-120h at 250rpm and 30°C. The L-asparaginase before and after the fusion glucoamylase catalytic region was expressed and purified by a nickel column. The recombinant bacteria fermentation broth was centrifuged at 10,000 rpm for 10 min to separate the bacteria from the fermentation supernatant, and the supernatant was collected and purified through a 0.22 μm filter membrane. Sample by Ni 2+ Purified by affinity chromatography column (GE Histrap FF 5mL) to obtain recombinant L-asparaginase.
[0047] Take 10 μL of the purified sample and perform SDS-PAGE electrophoresis. The protein band without the catalytic region of saccharification enzyme is about 42kDa in size, and a band of about 55kDa appears after fusion. Use NEB's deglycosylase EndoH (29kDa) to digest and recover the fused sample band figure 2 Afte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com