Engineering strain for co-displaying glucose oxidase and catalase on spore surface and application of engineering strain
A glucose oxidase and surface co-display technology, applied in the biological field, can solve the problems of low enzyme activity, low yield, complex detection methods, etc., and achieve the effects of wide pH adaptation range, improved stability, and changes in enzymatic properties.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
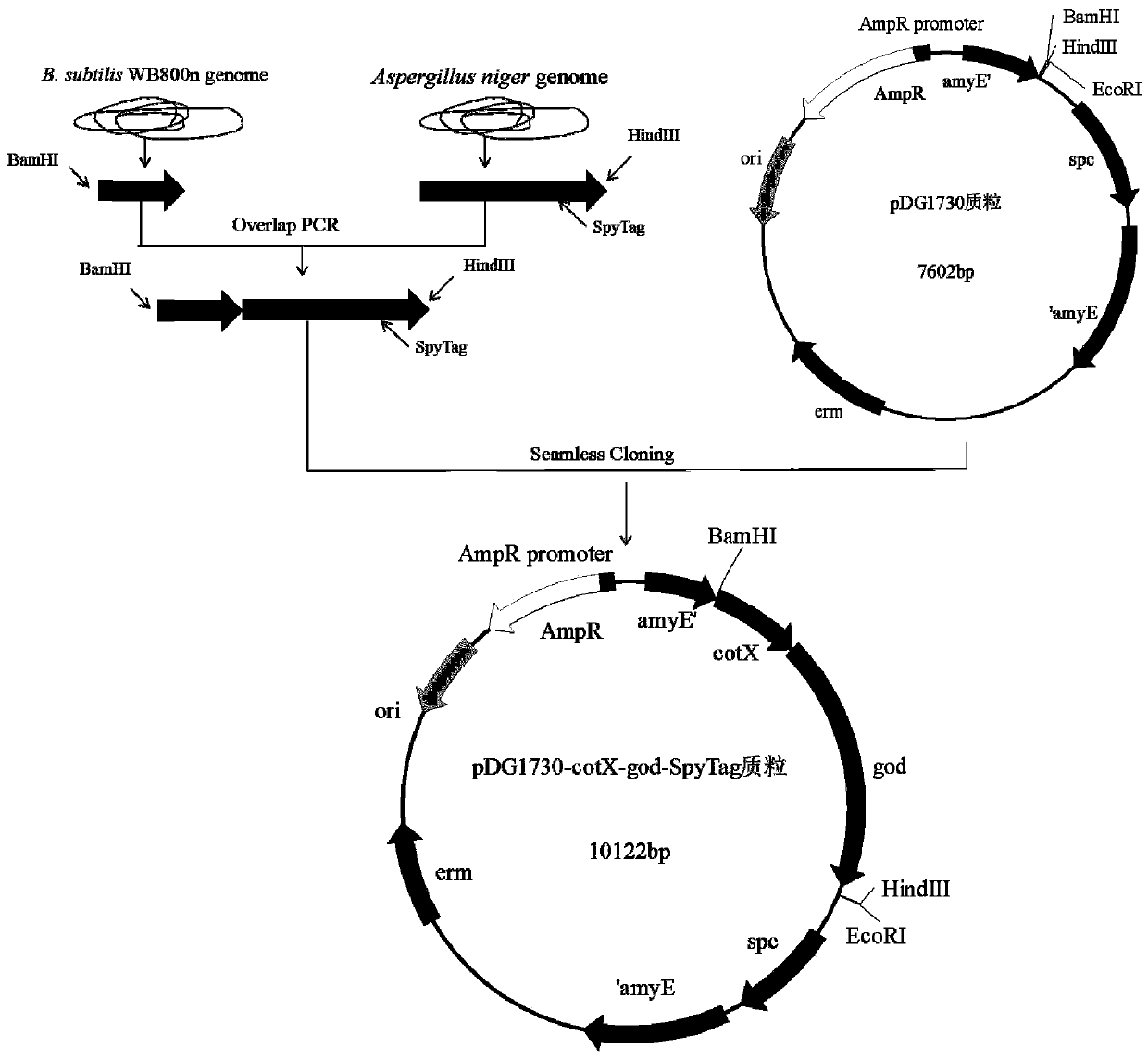
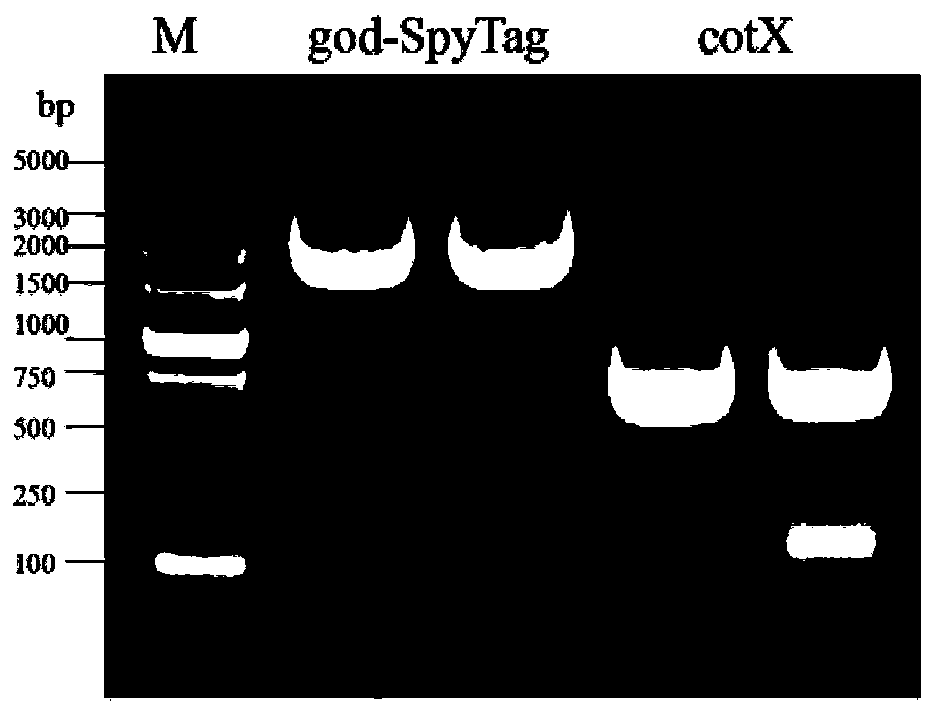
[0057] Example 1: Construction of recombinant plasmids
[0058] (1) Cloning to obtain the spore capsid protein gene fragment
[0059] Using the Bacillus subtilis WB800n genome as a template, primers were designed to amplify the gene fragment of cotX spore capsid protein and the gene fragment of P43 promoter by PCR.
[0060] The PCR primers of the CotX protein gene sequence are as follows:
[0061] cotX-F: 5'-aaaactggtctgatcggatccGATCAGACAAAGACGGAAAAACG-3'
[0062] cotX-R: 5'-gtctgcatGAGGACAAGAGTGATAACTAGGATGG-3'
[0063] The PCR reaction system is as follows:
[0064] 2×Phanta Max Master Mix 25μL, 10μmol / L upstream primer cotX-F 2.5μL, 10μmol / L downstream primer cotC-X 2.5μL, template 2.5μL, use ddH 2 O supplemented to 50 μL;
[0065] The above-mentioned PCR reaction is carried out according to the following procedures:
[0066] Pre-denaturation at 95°C for 3min; denaturation at 95°C for 15s, annealing at 60°C for 15s, extension at 72°C for 15s, 30 cycles; extension at 7...
Embodiment 2
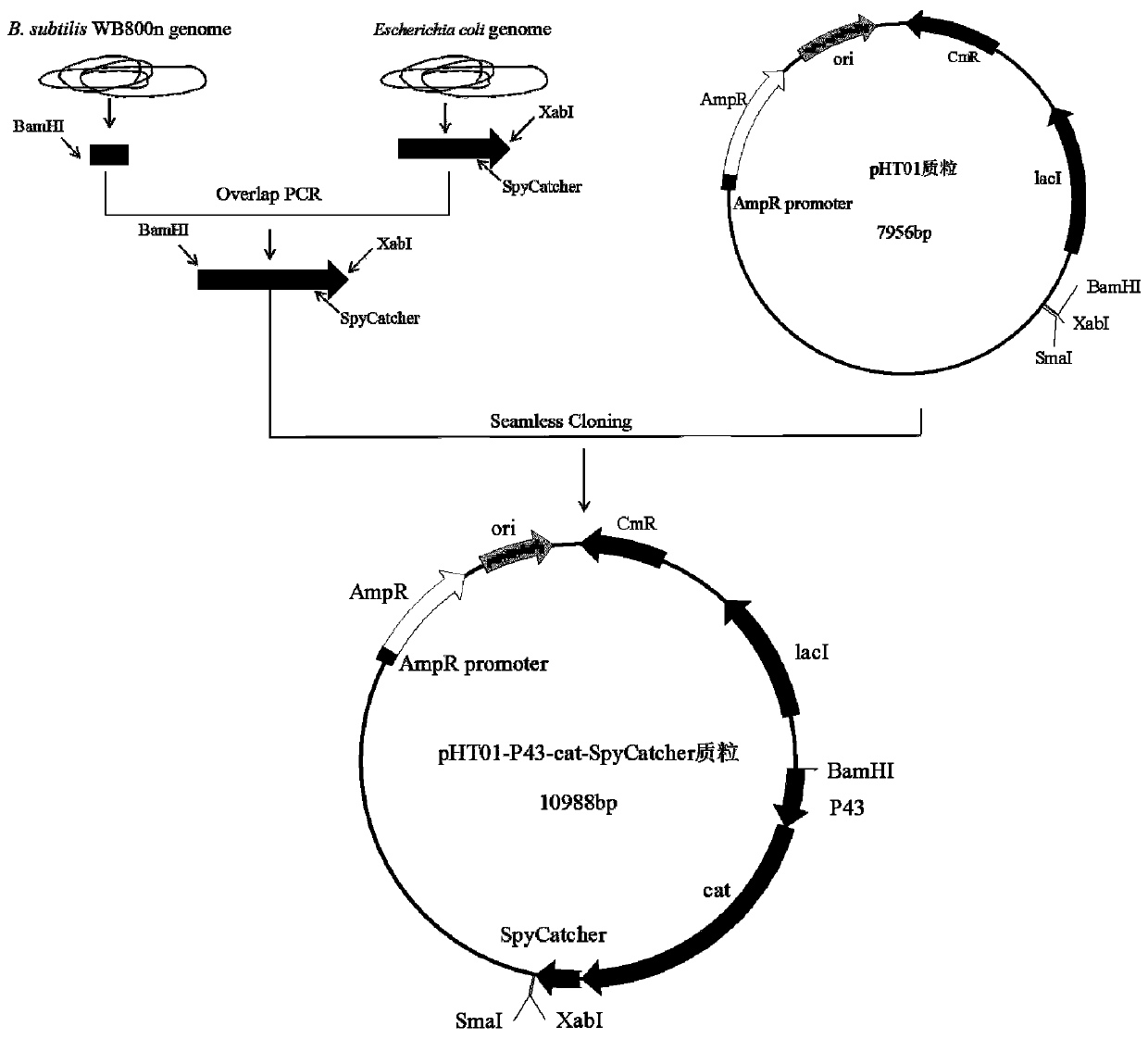
[0147] Example 2: Preparation of Bacillus subtilis WB800n electroporation competent cells
[0148] Pick a single colony of Bacillus subtilis WB800n on the surface of fresh LB solid medium and culture it overnight in 5 mL LB medium. Take 1 mL of the overnight culture and insert it into 50 mL of GM medium (LB+0.5M sorbitol), and culture it with shaking at 37°C until OD 600 is 1.0. The bacterial solution was placed in an ice-water bath for 10 minutes, centrifuged at 5000 rpm at 4°C for 8 minutes, and the bacterial cells were collected. The cells were resuspended with 20 mL of pre-cooled ETM medium (0.5M sorbitol+0.5M mannitol+10% glycerol), centrifuged at 5000rpm at 4°C for 8min, the supernatant was removed, and washed three times in this way. Resuspend the washed bacteria in 500 μL of ETM medium, aliquot them into EP tubes, and aliquot 60 μL in each tube.
Embodiment 3
[0149] Example 3: Transferring recombinant plasmid pDG1730-cotX-god-SpyTag into Bacillus subtilis WB800n
[0150] Add 5 μL of the recombinant plasmid pDG1730-cotX-god-SpyTag to 50 μL of competent cells WB800n, incubate on ice for 5 min, add to a pre-cooled electroporation cuvette (2mm), and perform electroporation at 2500V and 5ms. Immediately after the electric shock, add 1 mL of RM medium (LB + 0.5M sorbitol + 0.38M mannitol) preheated at 37°C to the electric transfer cup, revive and culture at 37°C for 3 hours, and spread on the LB plate containing spectinomycin . Inverted culture at 37°C, and strains resistant to spectinomycin were screened.
PUM
| Property | Measurement | Unit |
|---|---|---|
| thermal stability | aaaaa | aaaaa |
| thermal stability | aaaaa | aaaaa |
| thermal stability | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com