Methods for protein tyrosine phosphorylation profiling with variant sh2 domains
A technology of tyrosine phosphorylation and tyrosine phosphatase, which is applied in chemical instruments and methods, preparation of test samples, and general structural detail detection of gas analyzers, etc., can solve the problem that the Tyr phosphorylated proteome is far from complete. , covering complex issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
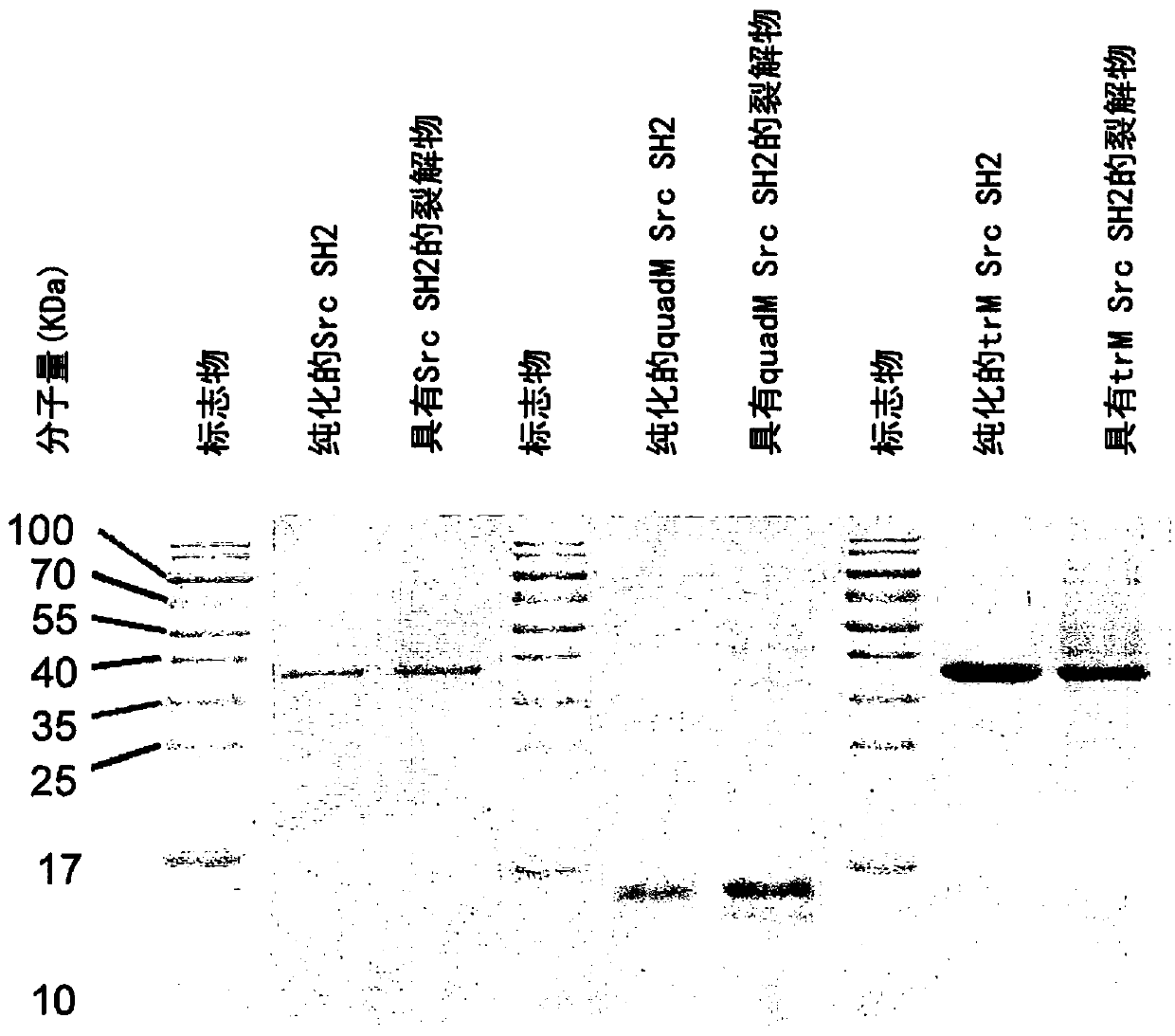
[0179] Example 1 - Expression and purification of wild-type and variant Src SH2 domains
[0180] Using standard techniques in the art, prepare the His-encoding DNA in a bacterial expression vector 6 / GST-tag tagged human SrcSH2 (residues Aspl44-Lys252, SEQ ID NO: 10), His 6 / GST-tag tagged TrM human Src SH2 (SEQ ID NO: 11) or His 6 - DNA sequence of tagged QuadM human Src SH2 (SEQ ID NO: 13).
[0181] Wild type and variant SH2 variants were expressed in E. coli BL21(DE3). Protein expression was induced with 0.5 mM IPTG overnight at 18°C. The cell pellet was resuspended in lysis buffer containing 2% Triton X-100, 1 mg / mL lysozyme, 3 μL totipotent nuclease in phosphate buffered saline (PBS) solution (pH 7.0) (benzonase) and 20mM imidazole, and sonicated at 400W for 180 seconds. Bacterial lysates were cleaned by centrifugation at 25,000g for 30 minutes and the supernatant obtained was used immediately or aliquoted and stored at -80°C for future use. Ni to be purchased from ...
Embodiment 2
[0183] Example 2 - Variant SH2 domains are better affinity reagents for pTyr-containing peptides from Jurkat cells than anti-pTyr antibodies on a mole-by-mole basis
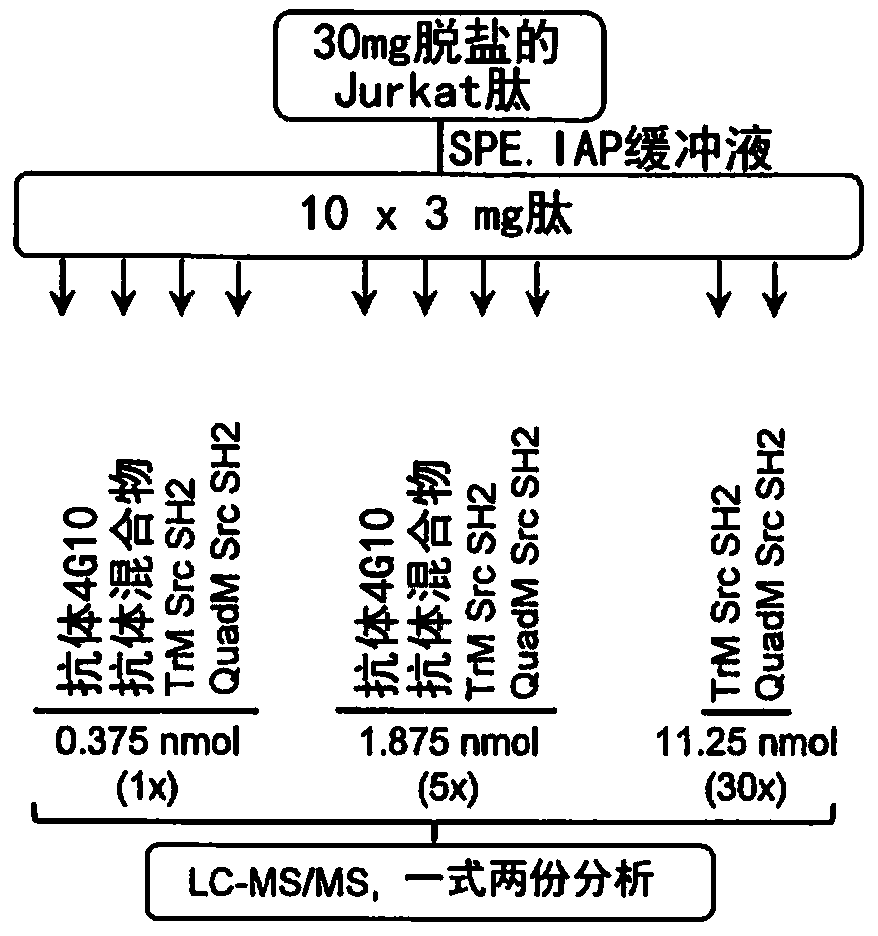
[0184] Experiments were performed to determine the relative potency of variant Src SH2 domains and commonly used anti-pTyr antibodies as pTyr-containing affinity reagents from biological samples. Assuming that each antibody molecule has two binding sites for its antigen, His was prepared in functionally equivalent molar amounts by using half the molar amount of antibody relative to the SH2 domain variant 6 / GST-tag tagged TrM Src SH2 domain (SEQ ID NO: 11) and His 6 - tagged QuadM Src SH2 domain (SEQ ID NO: 13) and anti-pTyr antibody.
[0185] The first experiment tested the relative affinity of the following affinity reagents for pTyr-containing peptides extracted from peptide mixtures prepared from Jurkat cells: His 6 / GST-tag tagged TrM Src SH2 (SEQ ID NO: 11), His 6 - Mixture of taggedQuadM Src SH2 (SEQ ID...
Embodiment 3
[0200] Example 3 - The sequence selectivity of TrM and QuadM Src SH2 domain variants becomes less pronounced when they are present at higher concentrations
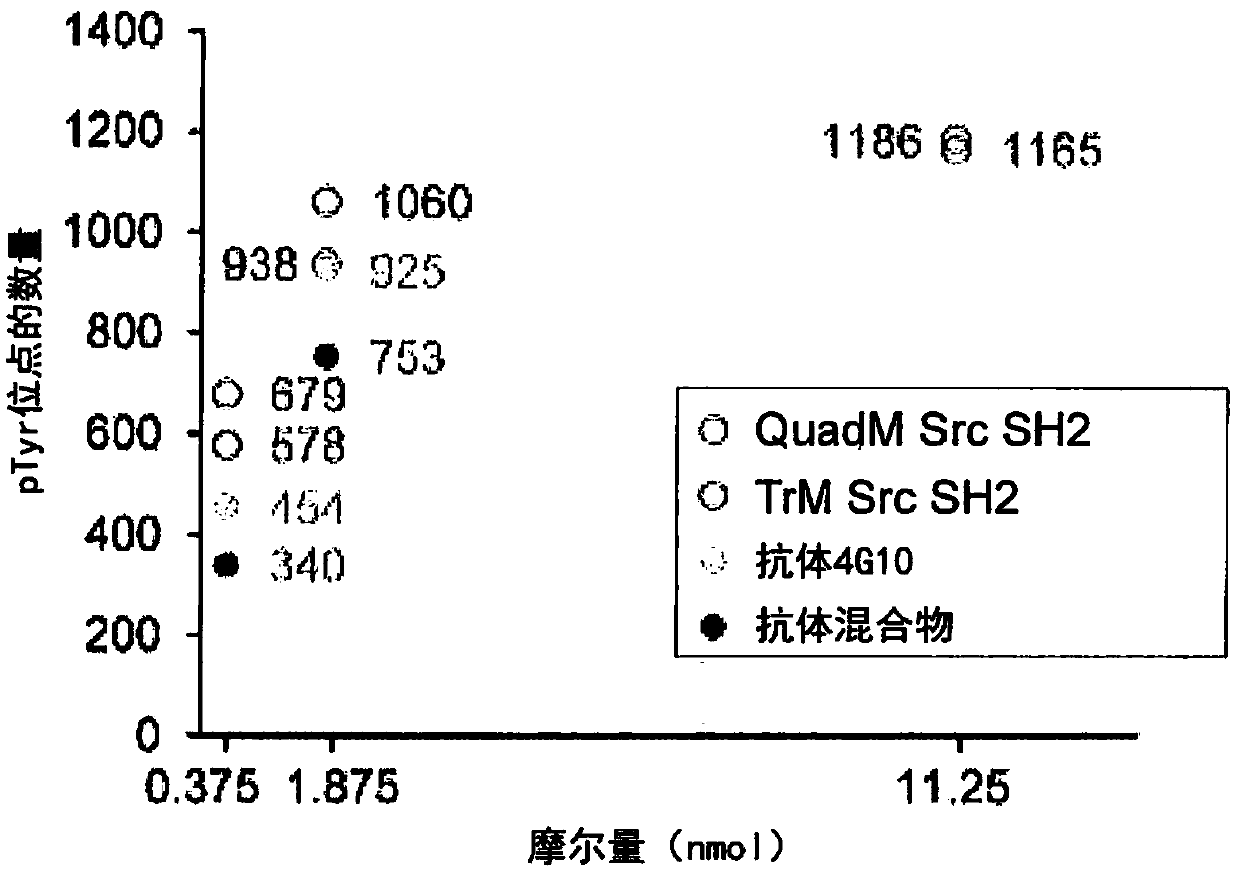
[0201] The pTyr-containing peptides identified in Example 2 were analyzed to calculate the selectivity distance between different affinity reagents for pTyr-containing peptides. Selectivity was measured by the distribution pattern of amino acid residues surrounding the pTyr residue in the identified phosphopeptides. The selectivity distance between two affinity reagents is measured by the Euclidean distance of the corresponding motifs.
[0202] image 3 The 1x amount between 4G10 (corner #1), antibody mix (corner #2), TrM Src SH2 (corner #3) and QuadM Src SH2 (corner #4) is shown (i.e., 0.375 nmol of pTyr binding sites) , selectivity distance for 5x amount (ie, 1.875 nmol of pTyr binding sites) and 30x amount (ie, 11.25 nmol of pTyr binding sites). The Euclidean distance between the sequences identified by the affinity...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com