Tumor marker STAMP-EP6 based on methylated modification
A methylation and tumor technology, applied in the direction of recombinant DNA technology, biochemical equipment and methods, microbial determination/testing, etc., can solve the problem of insufficient sensitivity and specificity, misdiagnosis, and affecting the sensitivity and accuracy of markers And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1, the nucleic acid sequence detected for STAMP-EP6
[0074] The sequence of the STAMP-EP6 tumor marker is provided, as shown in the following SEQ ID NO: 1 (chr2: 200327093-200327623 / hg19), wherein the underline indicates that the base is a methylated CpG site, and the number below the underline indicates the position of the site Numbering.
[0075]
[0076] The above sequence of SEQ ID NO:1 is treated with bisulfite and the sequence is as follows: SEQ ID NO:2 (wherein Y represents C or U):
[0077]
[0078]
[0079] The reverse complementary sequence of the nucleotide sequence shown in the above SEQ ID NO:1 is as follows: SEQ ID NO:3:
[0080] CG CCTAG CG CAGACT CG TTGCCCACCATCCTTCCCCCATCAGT CG TCCCTCTGAGG CG TATCTCTCTGCT CG GGC CG CAGGGGCTCATT CG CTTGGGGC CG GGAGGAGGACCTCCACAGTCTCTACCTC CG G CG GC CG A CG GCTTCCTTCCCTCTC CG CCCC CG CCCAGACCCCCTGAGCTGC CG GGGGAGCCAGGAGTCATTTATCTTTGCCTGG CGCG GGTTGGGATTTGTC CG GCTCTG...
Embodiment 2
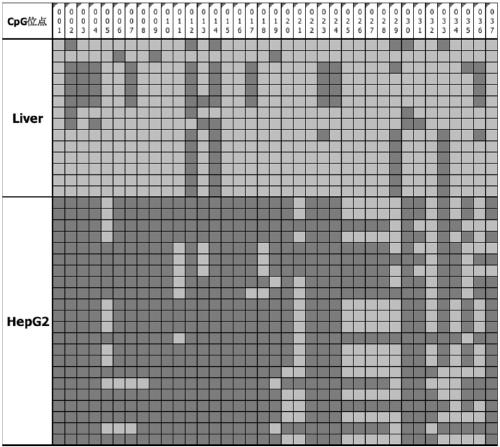
[0083] Example 2. Differences in the methylation of STAMP-EP6CpG sites between tumor cells and non-tumor cells—BSP-Bisulfite Sequencing PCR after bisulfite treatment
[0084] 1. Extract the genomic DNA of the liver cancer cell line HepG2 and the normal liver cell line;
[0085] 2. Treat the extracted HepG2 and normal liver cell line genomic DNA with bisulfite, respectively, as templates for subsequent PCR amplification;
[0086] 3. Design amplification primers (SEQ ID NO:5~6; Table 1) according to the sequence of SEQ ID NO:1, and routinely design primers to amplify;
[0087] 4. After PCR amplification, 2% agarose gel electrophoresis was used to detect the specificity of the PCR fragment, the gel was cut to recover the target fragment, ligated and inserted into the T vector, transformed into competent E. 10 clones were picked for Sanger sequencing.
[0088] Table 1, BSP primers
[0089]
[0090] The results of BSP verification of the methylation level of liver cancer cell...
Embodiment 3
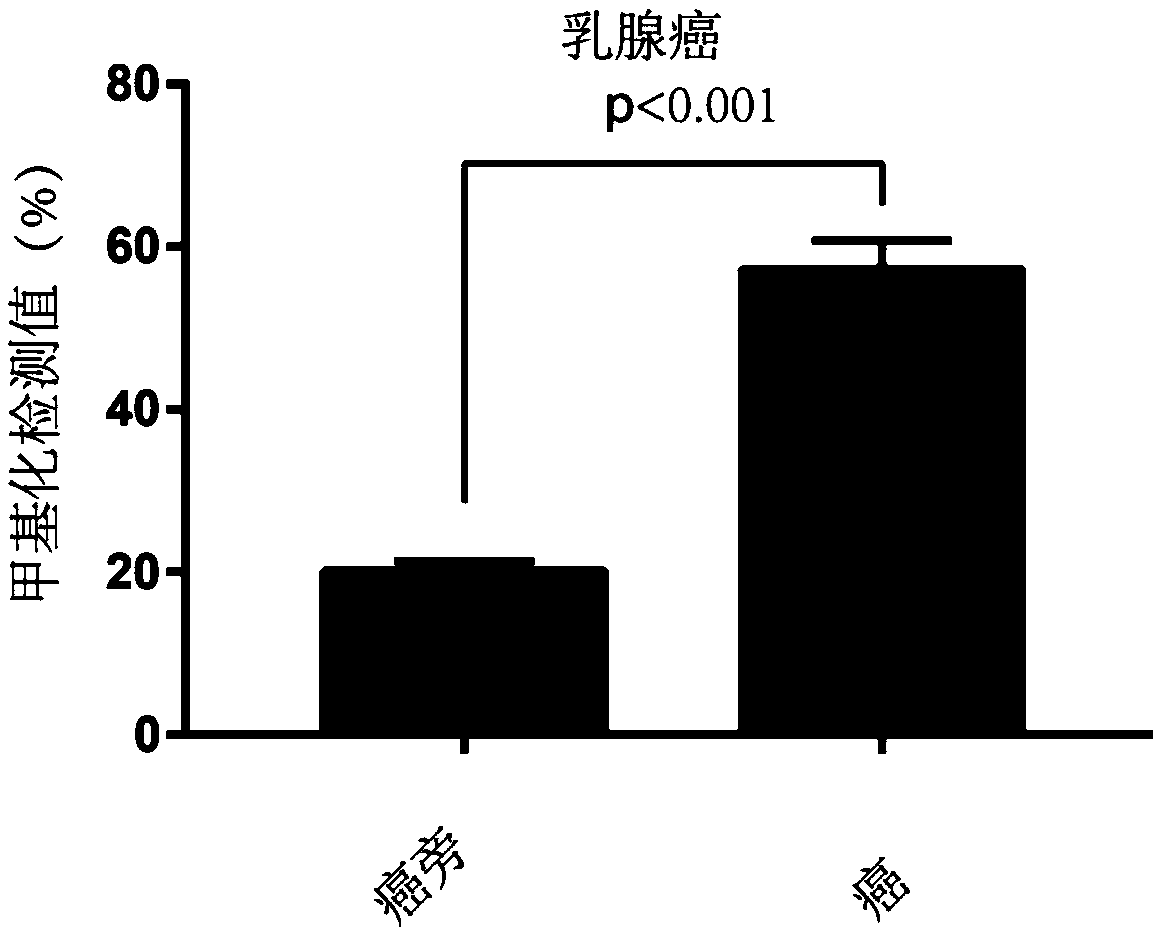
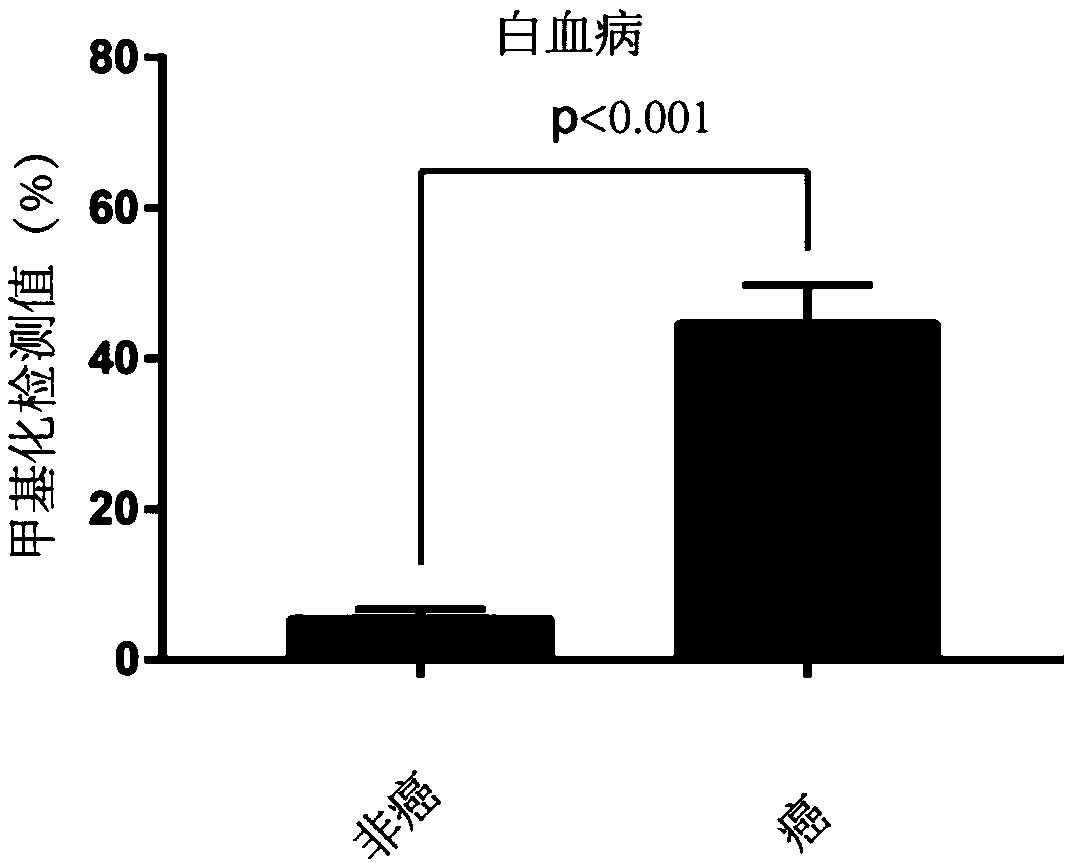
[0091] Example 3. Differences in methylation of STAMP-EP6 CpG sites between tumor cells and non-tumor cells—Pyrosequencing
[0092] 1. Obtain clinical samples: Obtain para-cancer / non-cancer-cancer tissue samples from the clinic, para-cancer / non-cancer samples are used as the control group, and cancer tissue samples are used as the tumor detection experimental group;
[0093] 2. DNA extraction: extract the DNA of the experimental group and the control group respectively; this experiment uses the phenol-chloroform extraction method, but is not limited to this method;
[0094] 3. Bisulfite treatment: treat the extracted DNA samples with bisulfite, and operate in strict accordance with the steps; in this experiment, EZ DNA Methylation-Gold Kit from ZYMO Research Company, Cat. No. D5006 was used, but not limited to this kit;
[0095] 4. Primer design: According to the characteristics of STAMP-EP6 sequence SEQ ID NO: 1, design PCR amplification primers and pyrosequencing primers to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com