Method and device for identifying and eliminating false positive in nucleic acid variation detection
A technology for mutation detection and nucleic acid mutation, applied in the fields of instrumentation, genomics, proteomics, etc., can solve problems such as affecting accuracy, increasing the workload of manual screening of false positive sites, and alignment errors in sequence comparison software, achieving The effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
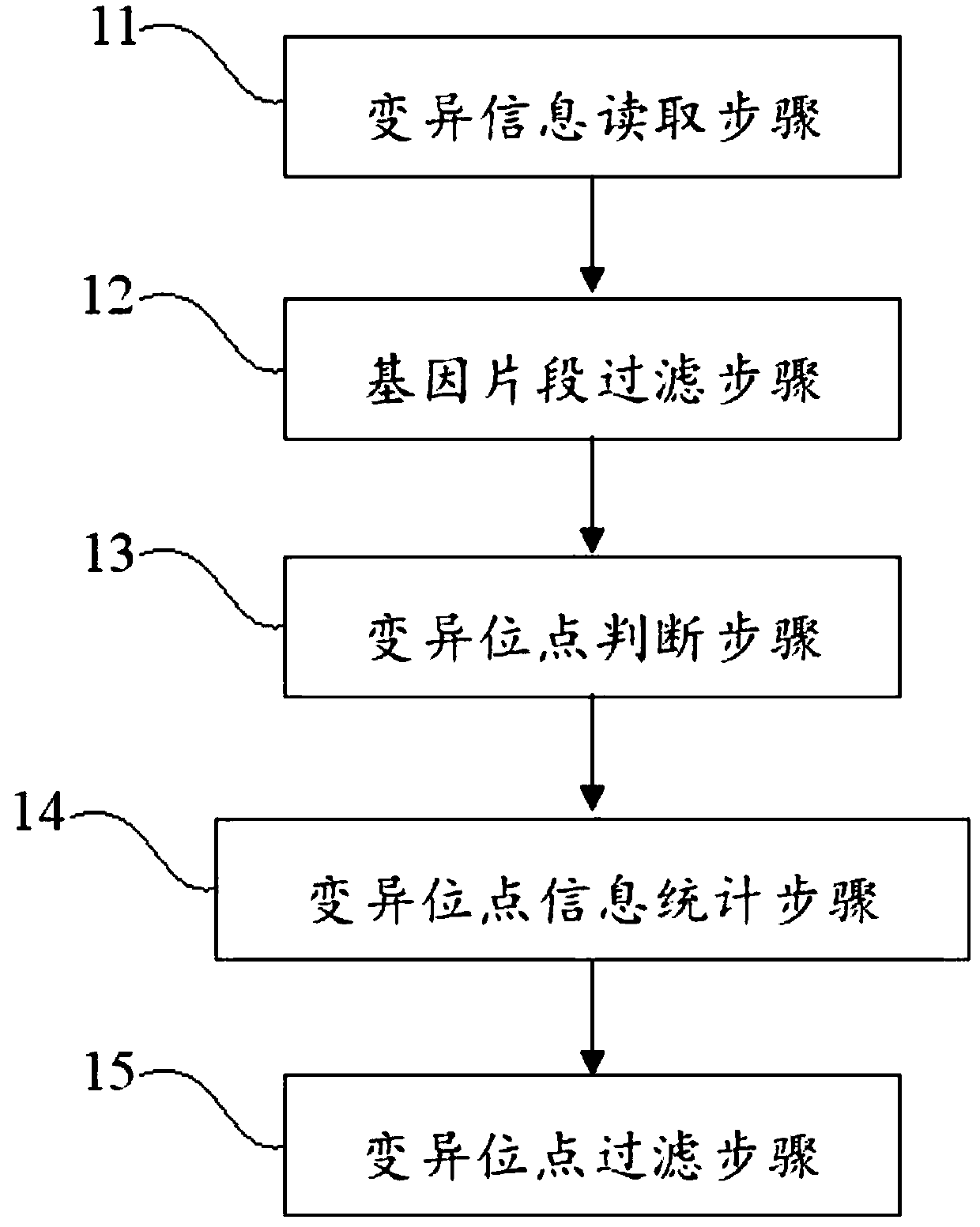
[0066] The method for identifying and eliminating false positives in nucleic acid variation detection in this example includes the following steps:
[0067] a, Step of reading mutation information: read the result file generated by the mutation detection software, the main information includes the mutation position, the base type at this position on the reference genome, and the variant base type at this position in the sample;
[0068] b, Gene fragment filtering step: Read the comparison file generated by comparing the off-machine sequence of the sample to be tested to the human reference gene, screen and obtain the read pair comparison results covered by each variant site, and then filter out the comparison results with the reference genome Compare read pairs with more than 2 mismatches, filter out read pairs with mutation base quality values less than 25, and filter out read pairs with inconsistent bases at mutation positions;
[0069] c, Judgment step of mutation site: j...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com