Kit utilizing stem loop primers to detect PIK3CA gene mutation sites
A stem-loop primer and mutation site technology, applied in the biological field, can solve the problems of cumbersome second-generation sequencing operations, limited detection capability, and high cost, and achieve the effects of no cross-reaction, high accuracy and sensitivity, and high precision.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Use the plasmid template containing the H1047R mutation site of the human PIK3CA gene (corresponding to each site to be tested), and use the primers and probes in Table 1 to perform qPCR to optimize and select the best stem-loop primers and MGB probes.
[0072] Wherein, the wild-type plasmids and mutant-type plasmids involved in the following tables can be prepared according to conventional plasmid construction methods and PCR cloning methods.
[0073] 1) Plasmid treatment and extraction:
[0074] Plasmid extraction was carried out using TIANGEN (HighPure Plasmid Kit, DP116) plasmid extraction kit, and the specific operation steps are detailed in the product manual. The extracted DNA was dissolved in Tris-HCl (10 mmol / L, pH 8.0), and the quality of the sample was detected and the concentration was determined by an ultraviolet spectrophotometer. Then, dilute the sample to 1000 copies / uL. Take 1 µL for ddPCR reaction.
[0075] 2) Prepare a PCR reaction system according...
Embodiment 2
[0092] Using the present invention to detect mutant cell line DNA samples, culturing cells containing the H1047R mutation site of the human PIK3CA gene, extracting DNA, using the specific stem-loop primers and MGB probes of the present invention to detect it, and at the same time, the minimum detection limit of mutation detection Determination.
[0093] Proceed as follows:
[0094] 1) Sample processing and DNA extraction:
[0095] Use a commercial DNA extraction kit to extract sample DNA, and refer to the kit instructions for specific operations.
[0096] Samples were diluted to 10ng / µL.
[0097] 2) Carry out PCR amplification according to the following amplification system (total volume 20 μL)
[0098]2 × ddPCR supermix for Probes 10μl
[0099] Each primer 0.005~1.0μmol
[0100] Probe 0.001~1.0μmol
[0101] 10ng / μL DNA template 0.1~1.0μL
[0102] Make up to 20μL with water;
[0103] Wherein, the primers in the above PCR amplification are the primers shown in SEQ ID NO...
Embodiment 3
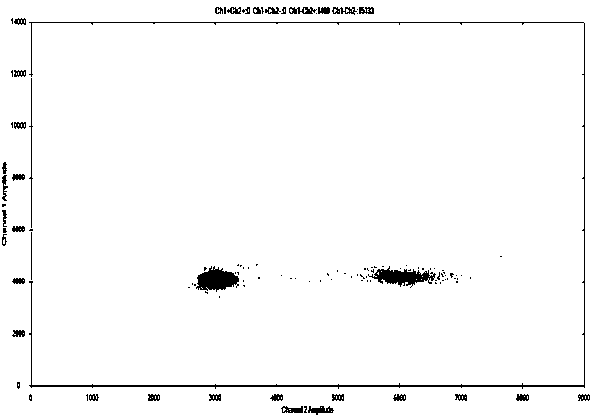
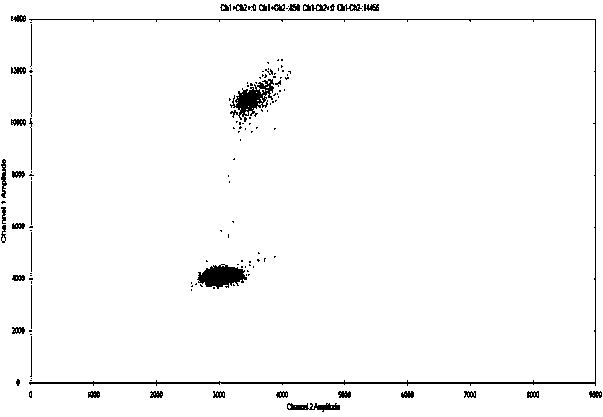
[0118] Using the stem-loop primers and common primers (Table 5) of the present invention to detect plasmid samples and mutant cell line DNA samples respectively, the sample acquisition and specific implementation steps are as in Example 1 and Example 2, and the obtained data are shown in Table 6.
[0119] Table 5 Common primer sequences
[0120]
[0121] Table 6 Data comparison between stem-loop primers and common primers
[0122]
[0123] The results show that the specificity and accuracy of the stem-loop primers of the present invention are better than those of common primers. As shown in Table 6, the positive detection rate of stem-loop primers used in the present invention can reach 100%, far exceeding the positive detection rate of common primers. The output rate has achieved unexpected technical effects. Stem-loop primers are designed so that bases in their stems are complementary to each other, and form a stem-loop structure by themselves without binding to the t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com