High-throughput method based on DNA (desoxyribonucleic acid) and RNA (ribonucleic acid) gene mutation detection
A high-throughput, DNA library technology, applied in biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of difficult probe design, affecting capture efficiency, and difficult analysis, so as to shorten the detection process and cycle , design simplification, good fusion detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
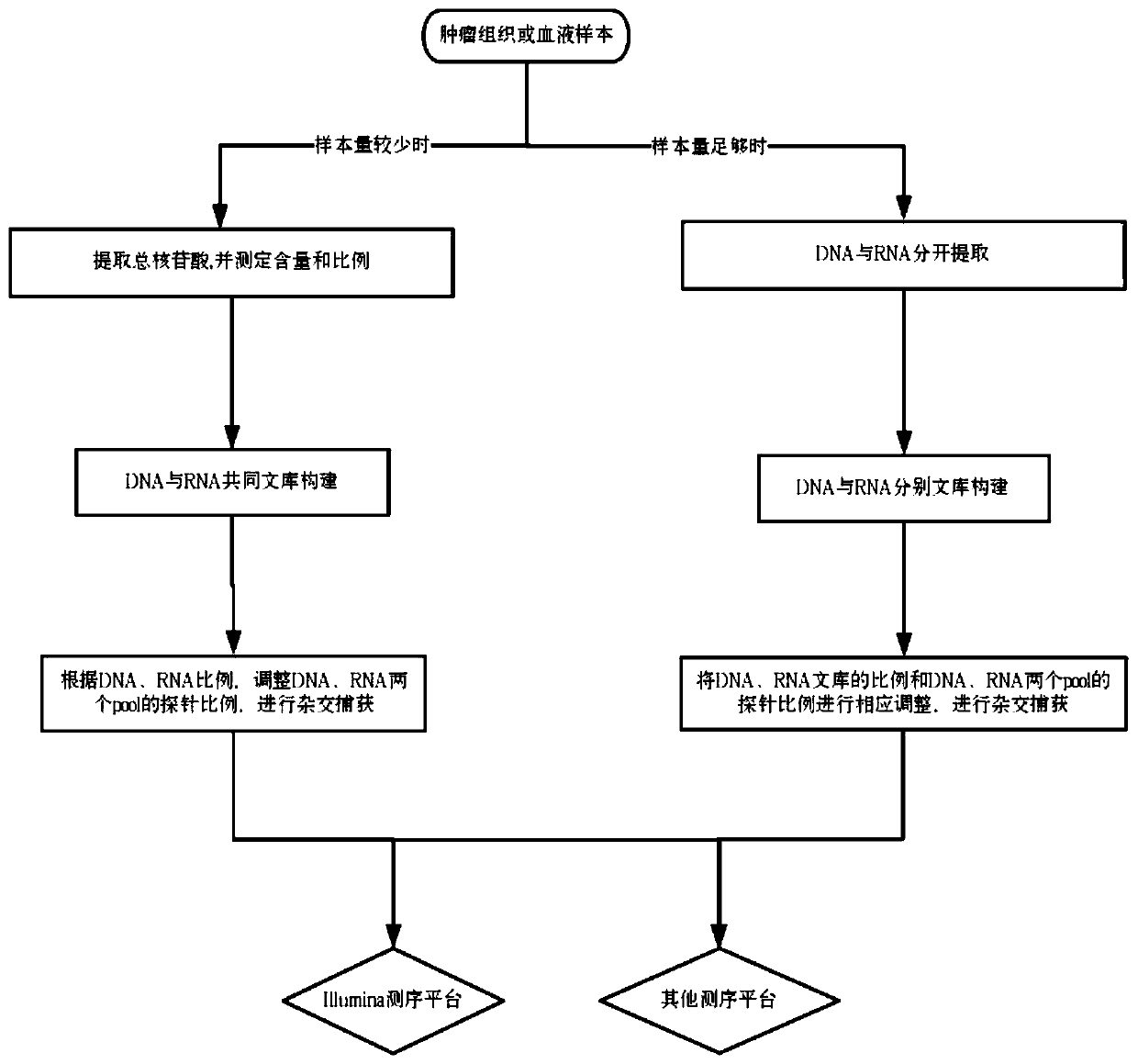
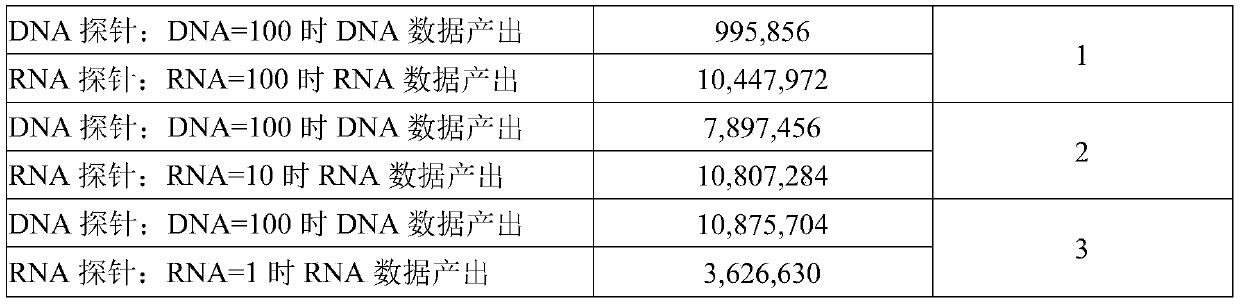
Image
Examples
Embodiment
[0068] This example illustrates the method of the present invention by taking gene detection of lung cancer targeted drugs and sequencing on the Illumina platform as an example.
[0069] Lung cancer is a common malignant tumor of the lung and one of the most threatening malignant tumors to human health and life. According to the latest statistics, there were about 733,000 new cases of lung cancer in my country in 2015, ranking first in the incidence of malignant tumors, and the incidence rate of men is about twice that of women. At the same time, the number of deaths from lung cancer in my country in 2015 was about 610,000, which also ranked first in the death rate of malignant tumors.
[0070] Scientific research has proved that cancer is caused by gene mutations in the human body. These gene mutations will cause cells to lose their normal regulatory ability, thereby proliferating indefinitely, and eventually leading to cancer. Targeted drugs can kill tumor cells carrying spec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com