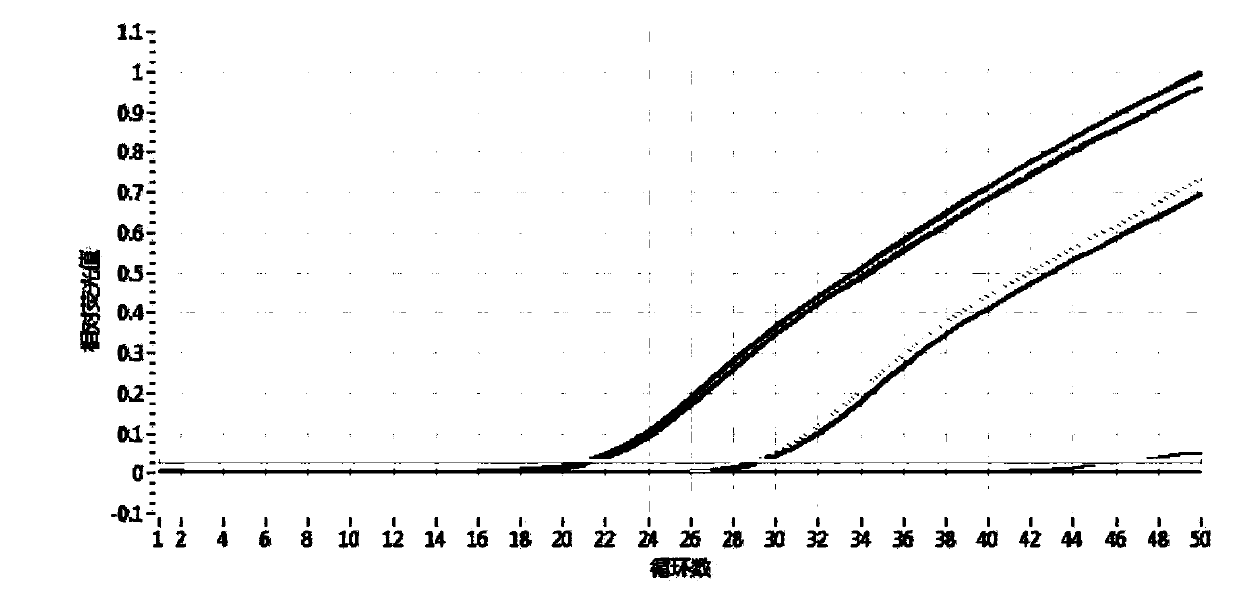
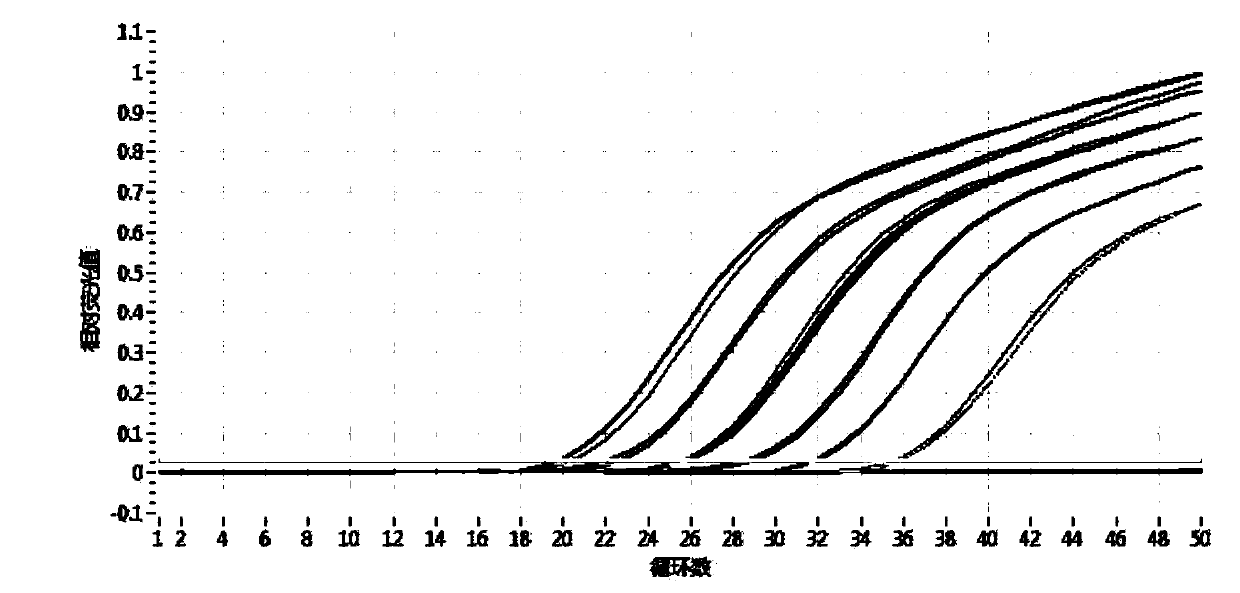
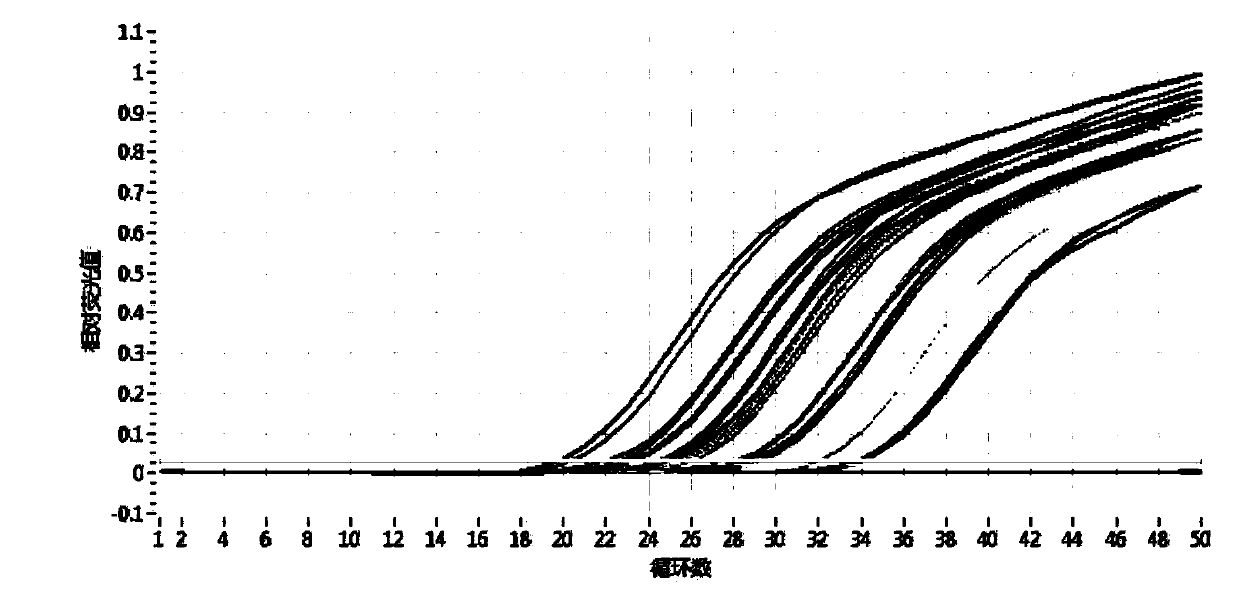
Method for identifying authenticity of mink fur products based on MGB probe
A technology of authenticity identification and mink, applied in the field of molecular biology, can solve the problems of high fluorescence background value and incomplete fluorescence quenching
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Detection of Suspected Mink Fur Products
[0034] 1. DNA extraction system
[0035] Take 10 mink guard hairs (average 1-2cm) and place them in a 2ml centrifuge tube, cut them into pieces, add 280μl TNE, 30μl SDS, 20μl DTT, 20μl proteinase K, add 150μl Buffer C-L (AXYGEN Animal Genome Extraction Kit), and immediately vortex Vortex for 1 min to mix well. After brief centrifugation, place the centrifuge tube in a 56°C water bath for overnight digestion. The centrifugation step was performed according to the instructions of the AXYGEN Animal Genome Extraction Kit. Add 350 μl of protein removal solution, vortex for 30 seconds to mix evenly, and centrifuge at 12000 r / min for 10 minutes. Place the DNA preparation tube in a 2ml centrifuge tube, transfer the centrifuged supernatant to the preparation tube, and centrifuge at 12000r / min for 1min. Discard the filtrate, add 500 μl washing solution, and centrifuge at 12000 r / min for 1 min. Discard the filtrate, add 700 μl desalte...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap