Fluorescence detection device for DNA (deoxyribonucleic acid) repeat sequences
A DNA repeat and fluorescence detection technology, applied in biochemical cleaning devices, enzymology/microbiology devices, and microbial determination/inspection, etc. The effect of low cost, convenient operation and simple structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0045] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below through specific embodiments in conjunction with the accompanying drawings.
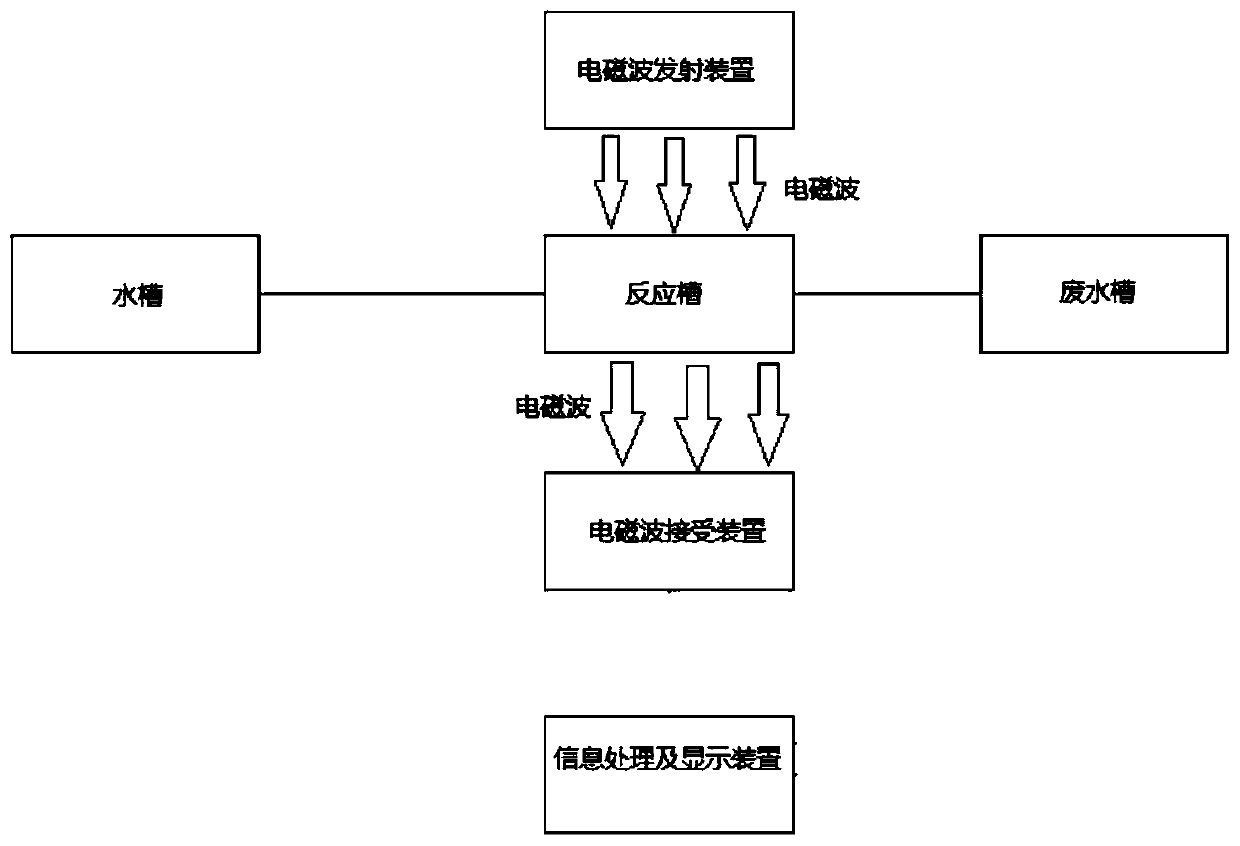
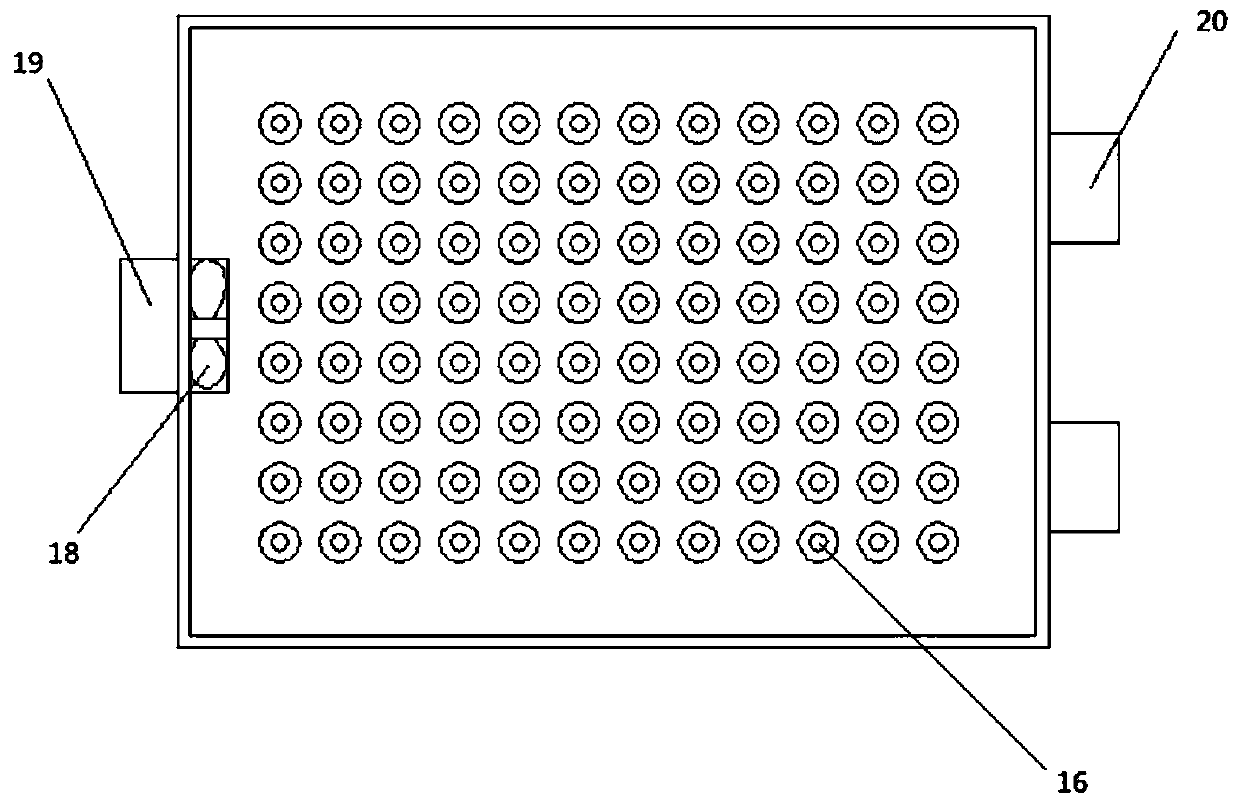
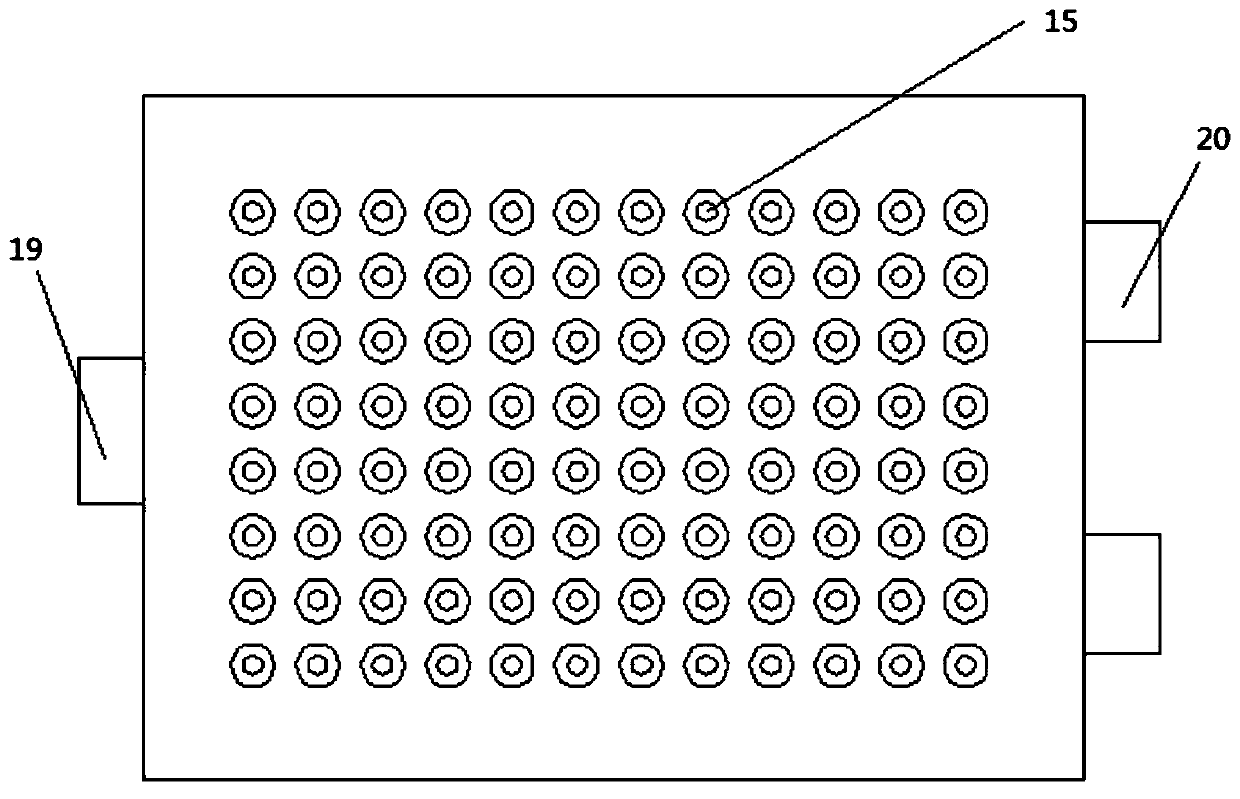
[0046] A fluorescence detection device in a DNA repeat sequence disclosed by the present invention includes a chute frame 1, a reaction tank, an electromagnetic wave emitting device 4, an electromagnetic wave receiving device 5, an information processing and display device, and a water tank. There are two water tanks, The water tank is provided with a heating device and a temperature control device, and the water tank is provided with a propeller 12, and the water tank is connected to the reaction tank. The electromagnetic wave emitting device 4 is located directly above the reaction tank, and the electromagnetic wave receiving device 5 is located directly below the reaction tank. Directly facing the electromagnetic wave emitting dev...
PUM
| Property | Measurement | Unit |
|---|---|---|
| emission peak | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com