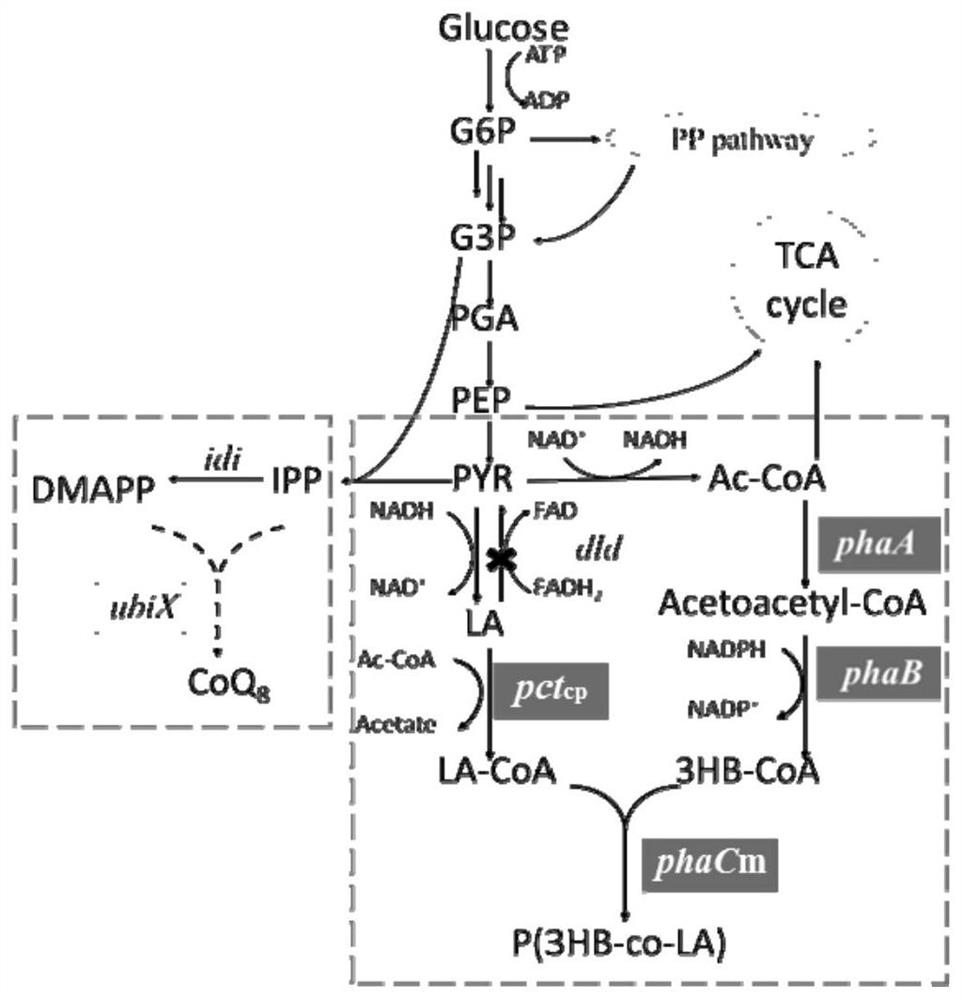
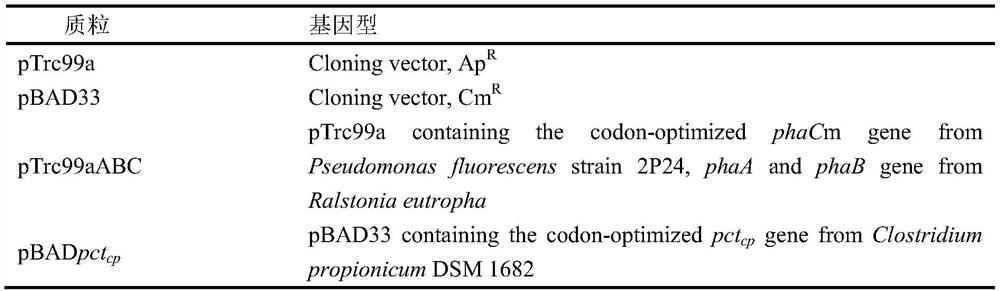
A method for increasing the content of lactic acid component in poly(3-hydroxybutyrate-co-lactic acid) synthesized by Escherichia coli
A technology of hydroxybutyric acid and Escherichia coli, applied in the field of metabolic engineering, can solve the problems of low cell productivity and slow growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1. Construction of recombinant Escherichia coli
[0035] Using MG1655 as the starting strain, red recombination technology is used to knock out the ubiX gene in the production process of coenzyme Q8. The specific operation is as follows:
[0036] The gene ubiX was knocked out by electroporation. The temperature-sensitive plasmid pKD46 is introduced into the host bacteria through calcium transduction to express exo, bet, and gam genes that assist homologous recombination, and are used to prepare electroporation competent. Using the primer ubiX-F with the sequence of SEQ ID NO: 1 and the primer ubiX-R with the sequence of SEQ ID NO: 2, the genome of the single-gene deletion bacteria containing the kanamycin resistance gene fragment was used as a template to PCR clone about 2000bp DNA fragments. The voltage of electroconversion is 1.8KV, and the current is 5ms. After transformation is completed, use kanamycin (kan) resistance to screen out recombinants, use the ...
Embodiment 2
[0046] Example 2.pct cp Constitutive promoters of genes optimize expression
[0047] Use the ldhA promoter derived from MG1655 to replace pBADpct cp Central original arabinose promoter. First use the primer PldhA-F with the sequence of SEQ ID NO:9 and the primer PldhA-R with the sequence of SEQ ID NO:10, use MG1655 as a template to PCR a gene of about 300bp containing the ldhA promoter, and use the sequence as SEQ ID Primer pBADpct of NO:11 cp -F and the sequence are primers pBADpct of SEQ ID NO: 12 cp -R to pBADpct cp The plasmid was used as a template for PCR amplification of the 6600bp gene except for the arabinose promoter. Use Clone MultiS One Step CloningKit for one-step connection. Use the primer PldhA-y-F with the sequence of SEQ ID NO:13 and the primer PldhA-y-R with the sequence of SEQ ID NO:14 to verify whether the replacement was successful or not by colony PCR. The strains with correct colony PCR were extracted plasmids and sent for sequencing, the sequen...
Embodiment 3
[0055] Example 3.M9 culture medium fermentation production polymer
[0056] Add 20g / L glucose to M9 medium, add 0.1mM IPTG to induce the expression of phaA, phaB and phaCm, add 50mM L-arabinose (L-arabinose) to induce pct cp gene expression. Incubate at 30°C for 48 hours to evaluate the performance of recombinant bacteria. The recombinant bacteria MG-01, JX04-01 and JX041-01 were used for fermentation. As shown in Table 7, compared with MG-01, the molar percentage of lactic acid component in P(3HB-co-LA) in the polymer produced by recombinant bacteria JX04-01 increased from 5.1% to 9.1%. An increase of 78.4%. And the polymer content is maintained above 75wt%. Using recombinant bacteria JX041-01 for fermentation, compared with MG-01 and JX04-01, the molar percentage of lactic acid component in P(3HB-co-LA) in the polymer further increased to 14.1%. Compared with MG-01, it increased by 176.4%, and compared with JX04-01, it increased by 54.9%, and the polymer content reached...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com