SNP molecular marker combination for establishing Luxi cattle pedigree
A molecular marker, Luxi cattle technology, applied in the field of molecular genetics, can solve the problems of lack of pedigree records, difficulty of quality traceability system, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
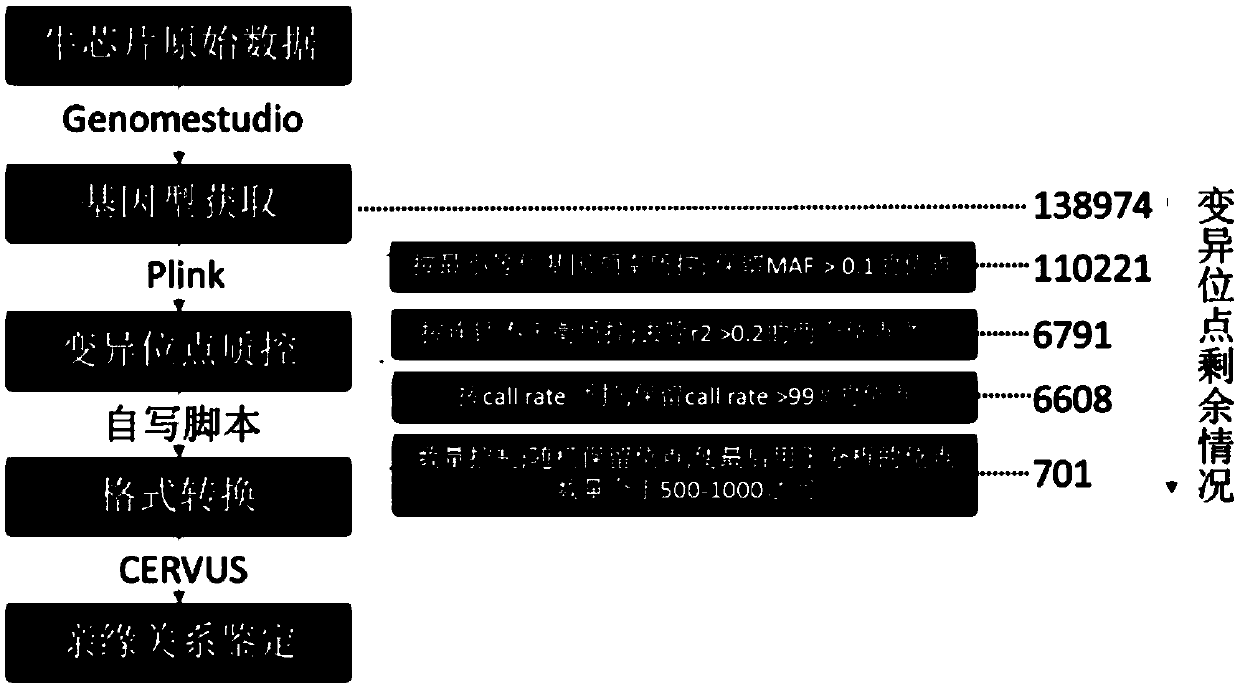
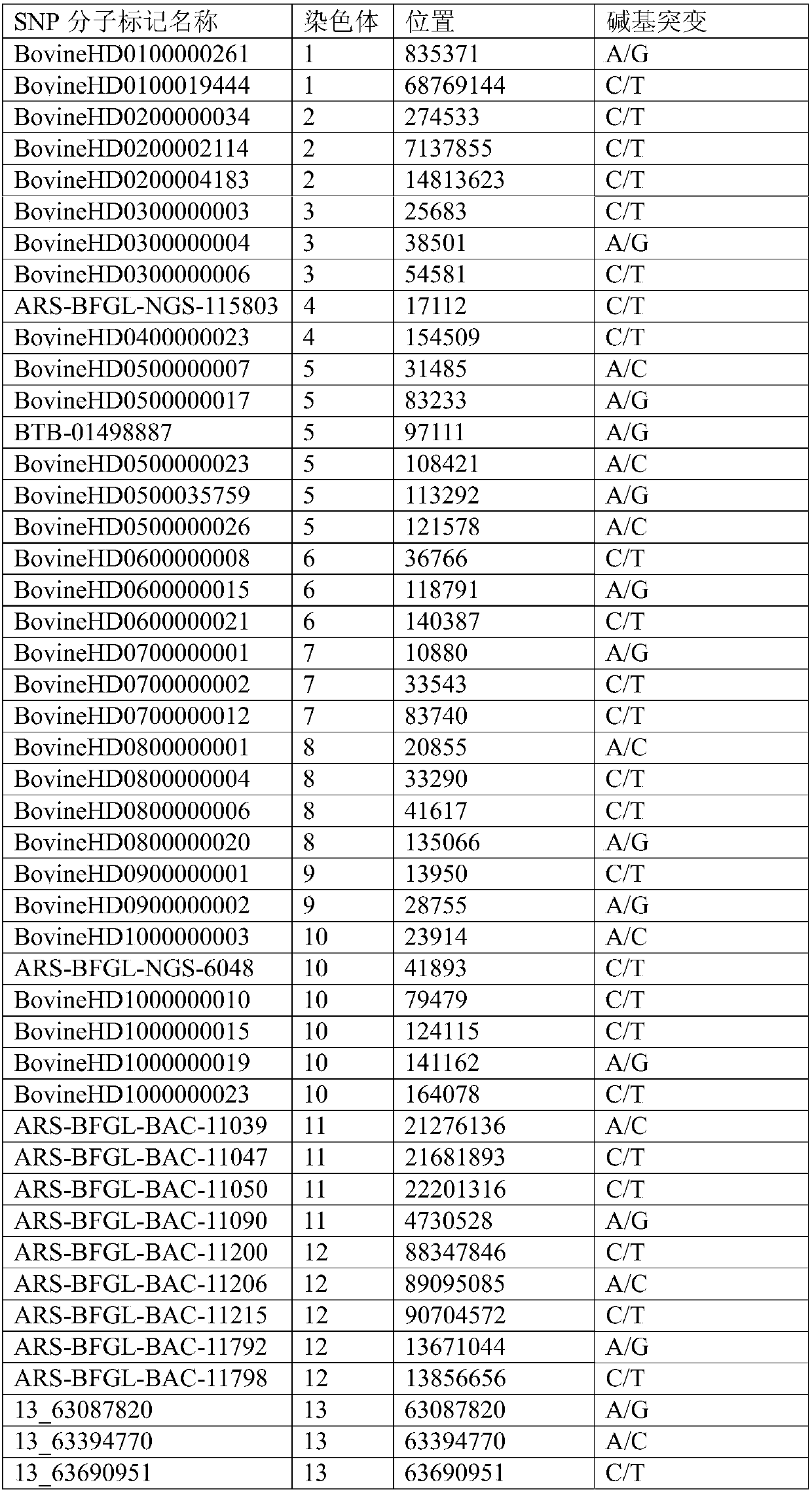
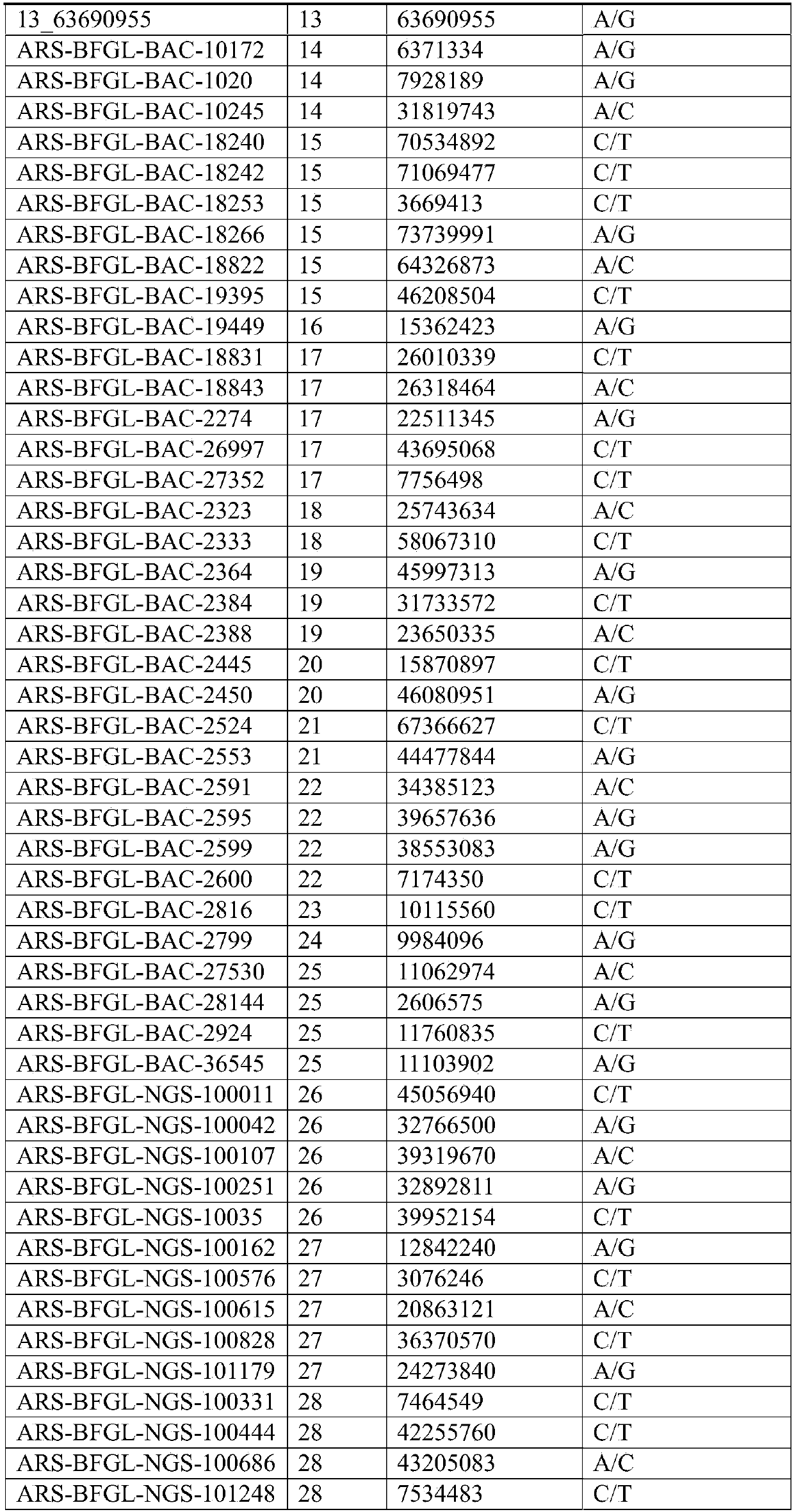
[0067] Example 1 Screening of 100 SNP molecular markers of Luxi cattle genetic relationship
[0068]Using the Bovine SNP150K Genotyping Bead Chip chip (purchased from Illumina) to analyze the results and data, the markers of XX Luxi cattle were genotyped. Refer to the US Department of Agriculture's screening criteria for SNP markers, and screen markers ① located on autosomes ② minimum allele frequency (MAF) greater than 0.35 ③ detection rate of each marker (Callrate) greater than 0.95 ④ similar on the same chromosome Neighbor distance greater than 8Mb; in order to avoid linkage disequilibrium among the selected SNP markers, separate chromosomes for screening. After preliminary screening, 701 polymorphic sites were obtained, and the number distribution of the sites on each chromosome is shown in Table 3.
[0069] chromosome SNPs average distance 1 49 7993223 2 38 7073627 3 39 8480873 4 40 7457215 5 30 7422463 6 29 6793028 7...
Embodiment 2
[0080] Example 2 Application of 100 Molecular Markers in Identification of Genetic Relationship of Luxi Cattle
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com