Fluorescent quantitative PCR detection method of human coronavirus and respiratory syncytial virus and application thereof
A human coronavirus, fluorescence quantitative technology, applied in the field of fluorescence quantitative PCR, to achieve good sensitivity and specificity, good reproducibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
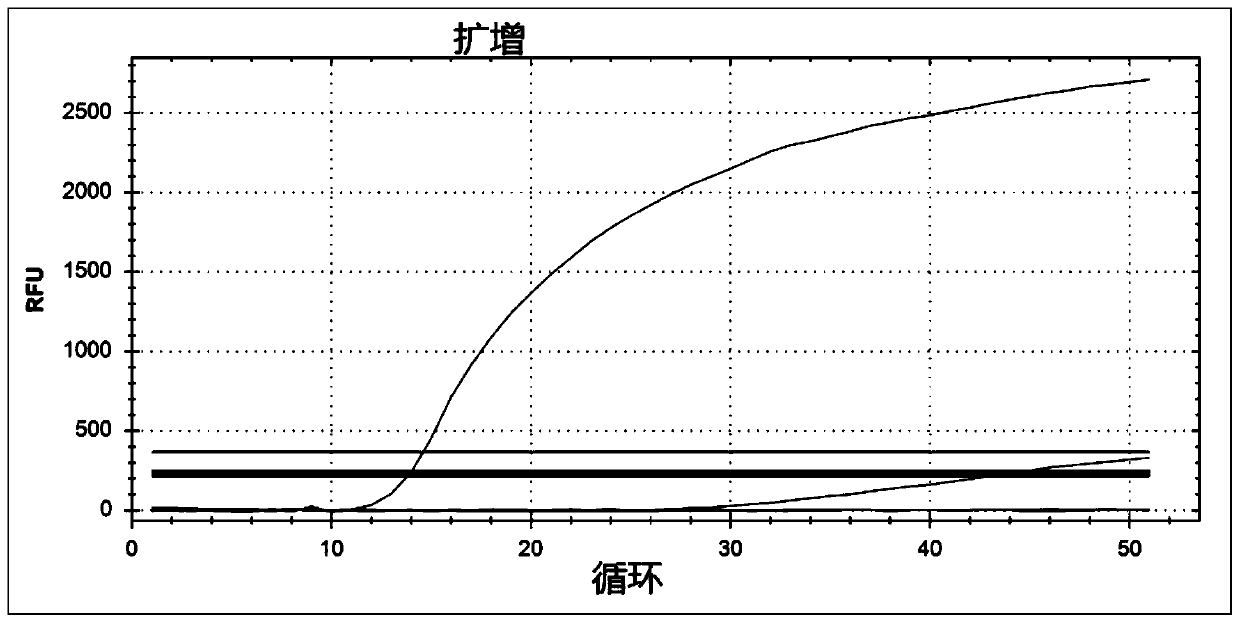
[0028] Embodiment 1 fluorescent quantitative PCR method detects human coronavirus OC43
[0029] 1.1 Extraction of nucleic acid
[0030] Take clinical samples from patients with acute respiratory infection, such as sputum, nasopharyngeal swabs, alveolar lavage fluid, etc., and extract nucleic acids according to conventional methods. The obtained nucleic acid was dissolved in 50 μl of eluent, and the extracted nucleic acid was quantified and tested for purity according to conventional methods. The extracted nucleic acid was divided into small portions and stored in a -80°C refrigerator.
[0031] 1.2 Design and synthesis of specific primers and probes
[0032] Using Primer 3 software, the fluorescent quantitative PCR forward primer and reverse primer were designed for the OC43 N protein gene sequence, and the probe was designed inside the gene. The design method is as follows: for the targeted target gene, TaqMan probes were designed using the sliding window method, the window...
Embodiment 2
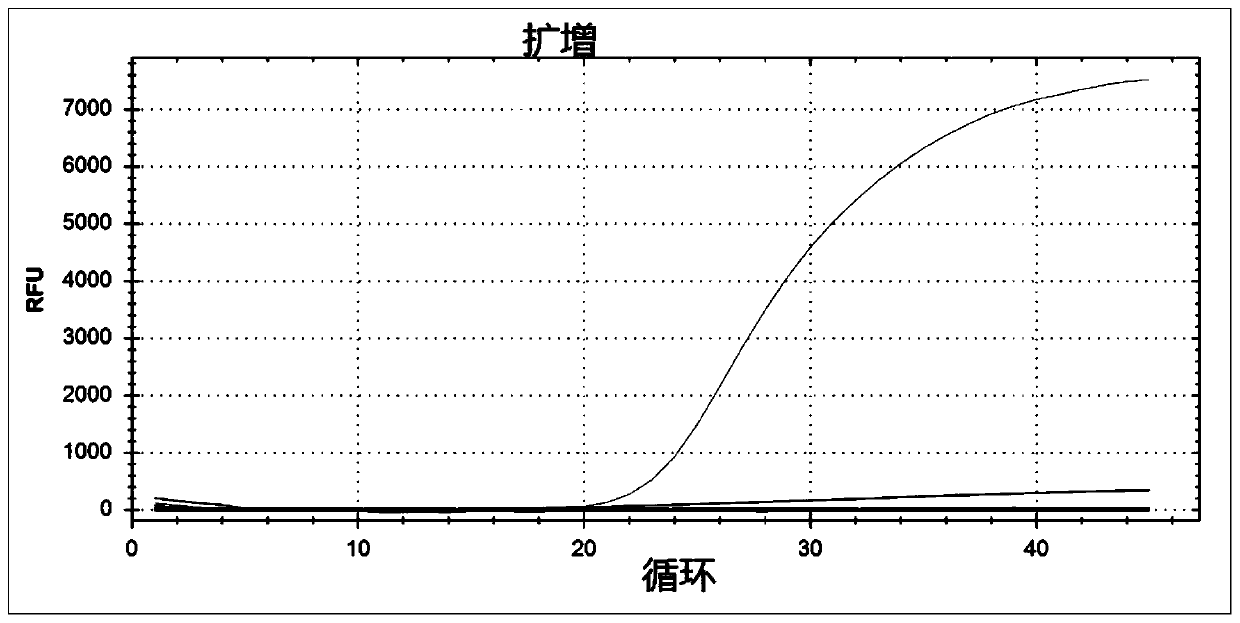
[0047] Embodiment 2 fluorescent quantitative PCR method detects human coronavirus HKU1
[0048] 2.1 Extraction of nucleic acid
[0049] Take clinical samples from patients with acute respiratory infection, such as sputum, nasopharyngeal swabs, alveolar lavage fluid, etc. Nucleic acid was extracted according to a conventional method, and the obtained nucleic acid was dissolved in 50 μl of eluent. Quantification and purity testing of the extracted nucleic acids were carried out according to conventional methods. The extracted nucleic acid was divided into small portions and stored in a -80°C refrigerator.
[0050] 2.2 Design and synthesis of specific primers and probes
[0051] Using Primer 3 software, the fluorescent quantitative PCR forward primer and reverse primer were designed for the HKU1 N protein gene sequence, and probes were designed inside the gene. The method is as described in 1.2. Primers and probes were synthesized according to conventional methods, and ROX f...
Embodiment 3
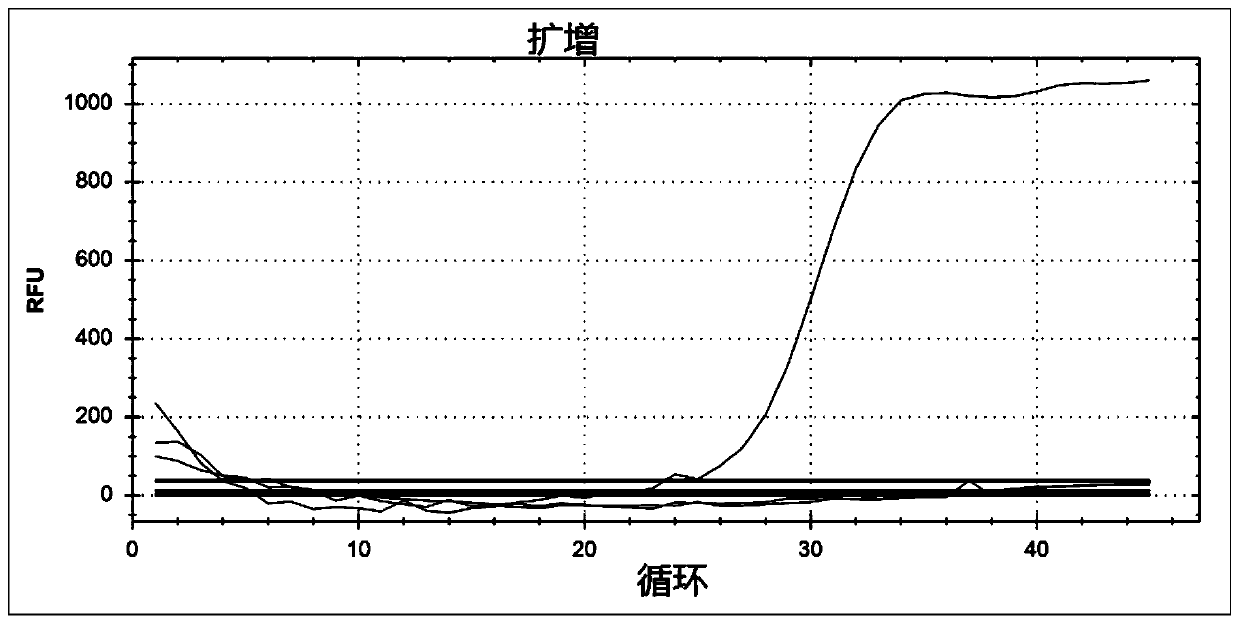
[0065] Embodiment 3 fluorescent quantitative PCR method detects respiratory syncytial virus RSVB
[0066] 3.1 Extraction of nucleic acid
[0067] Take clinical samples such as sputum, nasopharyngeal swabs, alveolar lavage fluid, etc. from patients with acute respiratory infection, extract nucleic acid according to conventional methods, and dissolve the obtained nucleic acid in 50 μl eluent. Quantification and purity testing of the extracted nucleic acids were carried out according to conventional methods. The extracted nucleic acid was divided into small portions and stored in a -80°C refrigerator.
[0068] 3.2 Design and synthesis of specific primers and probes
[0069] Using Primer 3 software, the forward primer and reverse primer of fluorescent quantitative PCR were designed for the gene sequence of RSVB N protein, and the probe was designed inside the gene. The method is as described in 1.2. Primers and probes were synthesized according to conventional methods, and a C...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com