Nucleobase editors comprising nucleic acid programmable DNA binding proteins
A technology for binding proteins and fusion proteins, used in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0650] Formulations of the pharmaceutical compositions described herein may be prepared by any method known or later developed in the art of pharmacology. Generally, such preparation methods comprise the steps of bringing into association one or more active ingredients with excipients and / or one or more other auxiliary ingredients, then, if necessary and / or desired, shaping the product and / or Or packaged as desired single-dose or multi-dose units.
[0651] The pharmaceutical formulation may additionally comprise a pharmaceutically acceptable excipient, which, as used herein, includes any and all solvents, dispersion media, diluents or other liquid vehicles, dispersion or suspension aids, surfactants, isotonic agents, Thickening or emulsifying agents, preservatives, solid binders, lubricants and the like, as appropriate for the particular dosage form desired. Remington's The Science and Practice of Pharmacy, 21 st Edition, A.R. Gennaro (Lippincott, Williams & Wilkins, Baltimo...
Embodiment 1
[0659] Embodiment 1: Cas9 deaminase fusion protein
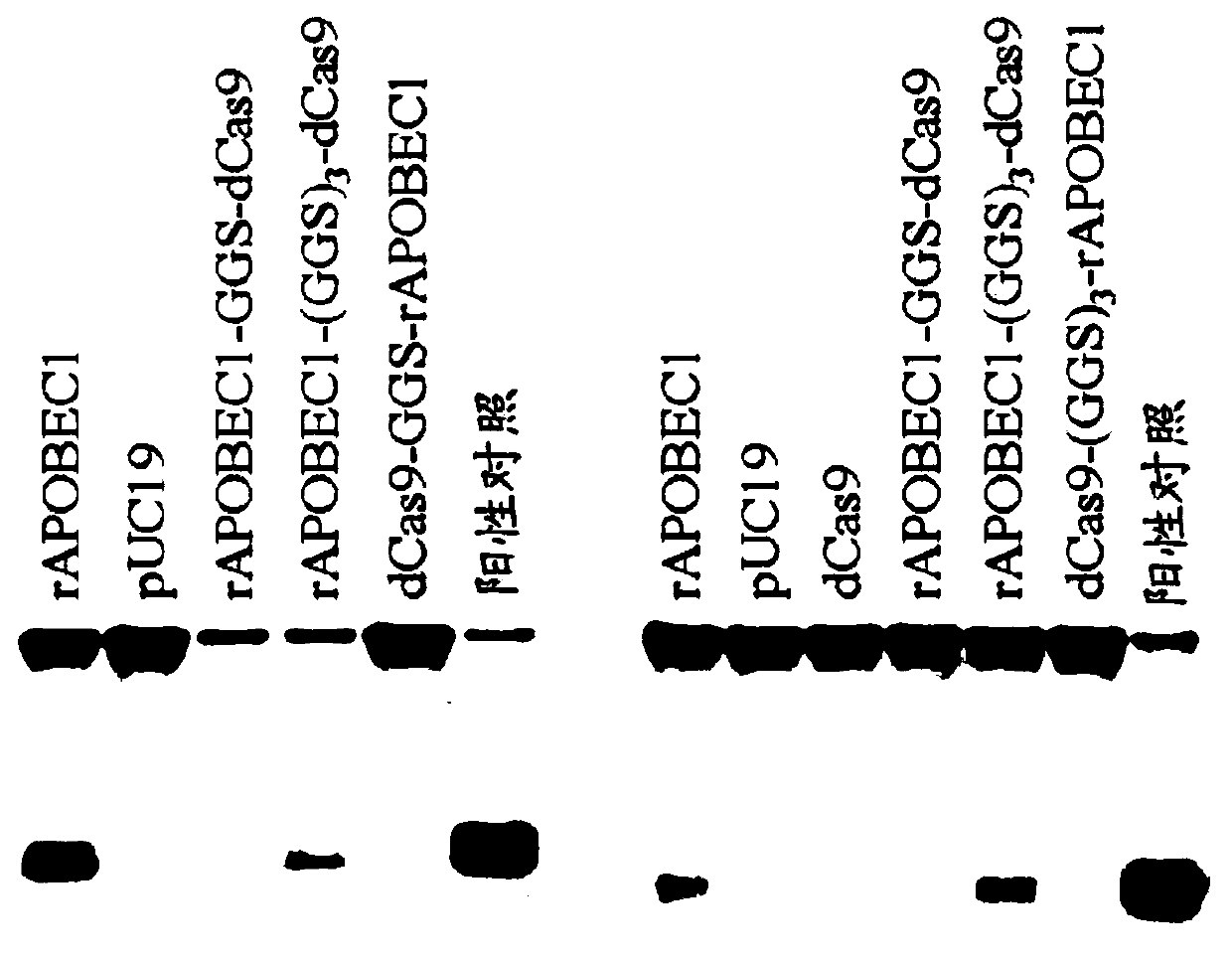
[0660] A number of Cas9:deaminase fusion proteins were generated and the resulting fusions were characterized for their deaminase activity. The following deaminases were tested:
[0661] Human AID (hAID):
[0662] MDSLLMNRRKFLYQFKNVRWAKGRRETYLCYVVKRRDSATSFSLDFGYLRNKNGCHVELLFLRYISDWDLDPGRCYRVTWFTSWSPCYDCARHVADFLRGNPYLSLRIFTARLYFCEDRKAEPEGLRRLHRAGVQIAIMTFKDYFYCWNTFVENHERTFKAWEGLHENSLSRAFQLLRILLLR(
[0663] Human AID-DC (hAID-DC, a truncated form of hAID with 7-fold increased activity):
[0664] MDSLLMNRRKFLYQFKNVRWAKGRRETYLCYVVKRRDSATSFSLDFGYLRNKNGCHVELLFLRYISDWDLDPGRCYRVTWFTSWSPCYDCARHVADFLRGNPNLSLRIFTARLYFCEDRKAEPEGLRRLHRAGVQIAIMTFKDYFYCWNTFVENHERTFKAWEGLHENSVRLSRQLR:(0) IDSLLM
[0665] Rat APOBEC1 (rAPOBEC1):
[0666] MSSETGPVAVDPTLRRRIEPHEFEVFFDPRELRKETCLLYEINWGGRHSIWRHTSQNTNKHVEVNFIEKFTTERYFCPNTRCSITWFLSWSPCGECSRAITEFLSRYPHVTLFIYIARLYHHADPRNRQGLRDLISSGVTIQIMTEQESGYCWRNFVNYSPSNEAHWPRYPHLWVRLYVLELYCIILGLPPCLNILRRKQPQL...
Embodiment 2
[0699] Example 2: Deamination of DNA Target Sequences
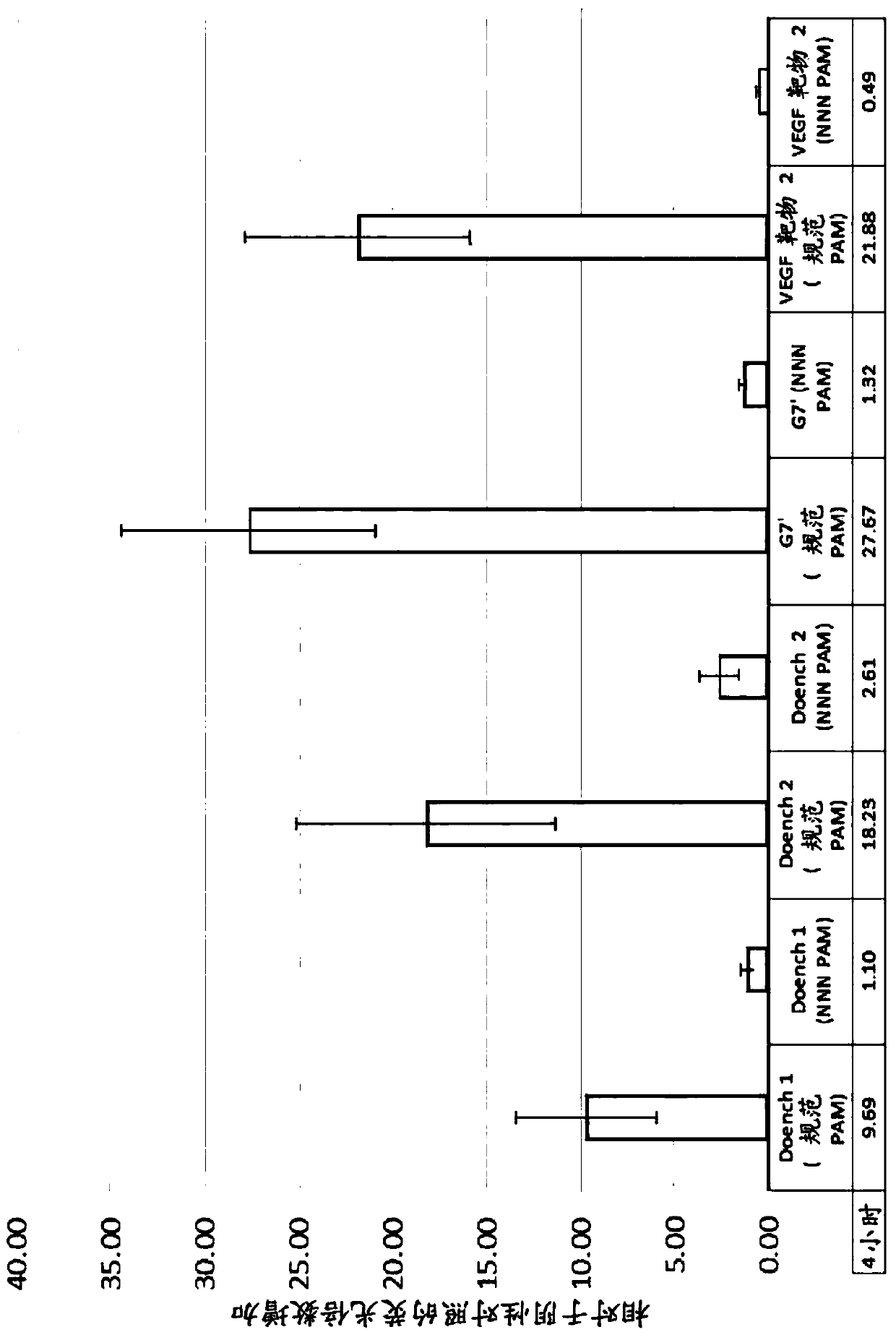
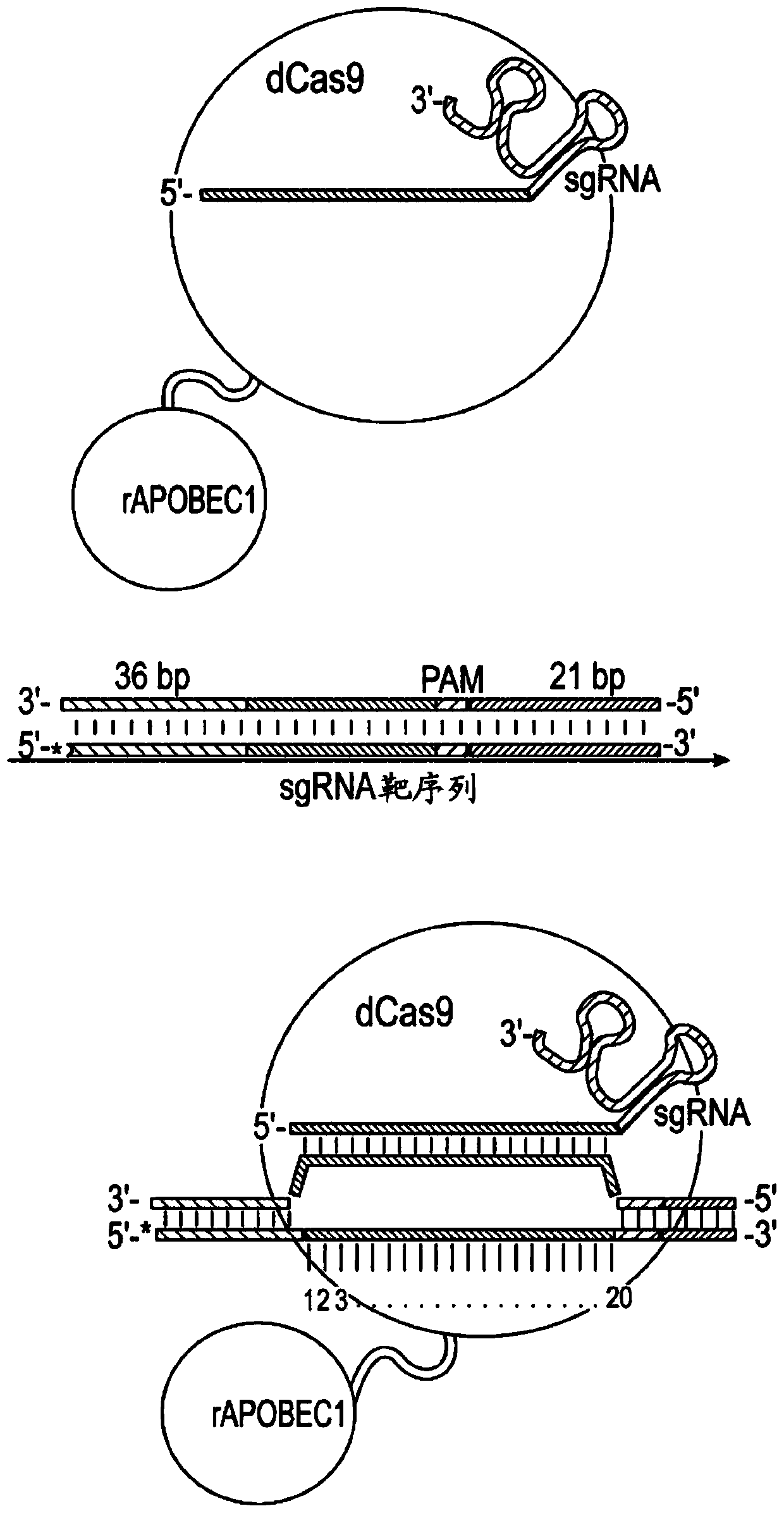
[0700] Exemplary deamination targets. The dCas9:deaminase fusion proteins described herein can be delivered to a cell in vitro or ex vivo or to a subject in vivo, and when the target nucleotide is in position 3-11 with respect to the PAM, can be used to achieve C to T or G to A transition. Exemplary deamination targets include, but are not limited to, the following: CCR5 truncation: Any codon encoding Q93, Q102, Q186, R225, W86, or Q261 of CCR5 can be deaminated to generate a stop codon, which results in CCR5 The non-functional truncation of , is used in HIV treatment. APOE4 Mutations: The mutant codons encoding the C11R and C57R mutant APOE4 proteins can be deaminated to revert to wild-type amino acids, which find application in the treatment of Alzheimer's disease. eGFP truncation: Any codon encoding Q158, Q184, Q185 can be deaminated to generate a stop codon, or the codon encoding M1 can be deaminated to encode I,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com