Primer composition and application thereof to identification of pheretima asiatica
A kind of primer composition, the technology of ginseng hairworm, applied in the direction of microbial determination/testing, recombinant DNA technology, biochemical equipment and method, etc., can solve the problem of being difficult to identify
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0055] 1. Reagents
[0056] Universal primers LCO1490 and HCO2198 and primers dl-F3, dl-FIP, dl-BIP, dl-B3 were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.; dNTP Mixture (i.e. dNTPs) (TaKaRa company); DEPC water (Biosharp ); Agarose (Lonza); Dehydrated alcohol (Tianjin Damao Chemical Reagent Factory); 6 × Loading Buffer (TaKaRa company); 10000 × SYBR GreenI nucleic acid dye (Beijing Soleibao Technology Co., Ltd.); DuRed nucleic acid dye ( Beijing Fanbo Biochemical Co., Ltd.); D2000 DNA Marker (Beijing Tiangen Biochemical Technology Co., Ltd.); TAE (50×) (Hefei Zhihong Biotechnology Co., Ltd.); Bst 2.0WarmStart DNA polymerase (New England Biotechnology Co., Ltd. , supporting reagents include 10× isothermal amplification reaction buffer); Tks Gflex DNA polymerase and 2×Gflex PCRBuffer (2×Gflex PCRBuffer contains dNTPs) (TaKaRa).
[0057] 2 Species identification of samples to be tested
[0058] 1. Acquisition of the COⅠDNA fragment of the sample to be tested
[...
Embodiment 2
[0074] Example 2 Investigation of LAMP Amplification Sensitivity
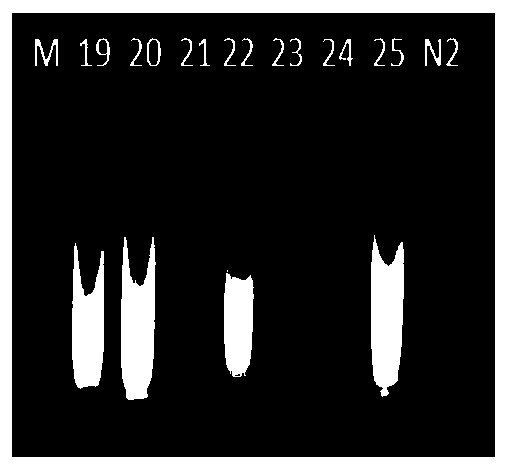
[0075] Select a sample that has been molecularly identified as genuine Dilong Ginseng (Sample No. 1) for sensitivity investigation. 10,000 dilutions are used as templates, amplified according to the LAMP amplification system and amplification conditions in Example 1, and detected by agarose gel electrophoresis. The electrophoresis results are as follows: Figure 7 shown. Among the figure, M represents marker (molecular weight marker), and labels 1-5 respectively represent that the templates in the amplification system are stock solution, 10-fold dilution, 100-fold dilution, 1000-fold dilution and 10000-fold dilution; from the results It can be seen that the template DNA can still amplify bands after being diluted 10000 times (concentration about 0.04 ng / μl), indicating that the primer composition of the present invention has higher sensitivity.
Embodiment 3
[0076] Example 3 Investigation of LAMP Amplification Reaction Time
[0077] Select a sample (sample number 1) that has been identified as a genuine product of Dilong ginseng (sample number 1) to investigate the amplification time. The LAMP amplification system and amplification conditions in Example 1 are used. The only difference is that the amplification time is set to 0 min. , 10min, 20min, 30min, 40min, 50min, 60min, and use agarose gel electrophoresis to detect the amplification results, the electrophoresis results are as follows Figure 8 shown. In the figure, M represents a marker (molecular weight marker), and the labels 1-7 represent the sample amplification results with a reaction time of 0-60 minutes respectively, and N is a negative control (DEPC water); it can be seen from the results that the amplification time Bands were produced when it reached 30 minutes, and the amplification effect was the best when the reaction was 50 minutes and 60 minutes.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com