Product for breast-cancer-II screening and evaluation based on saliva specific glycoprotein carbohydrate chain structure and application thereof
A breast cancer and glycoprotein technology, which can be used in measurement devices, instruments, scientific instruments, etc., can solve the problems of too many lectins, and the lack of simple and effective screening, diagnosis and evaluation of stage II breast cancer.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
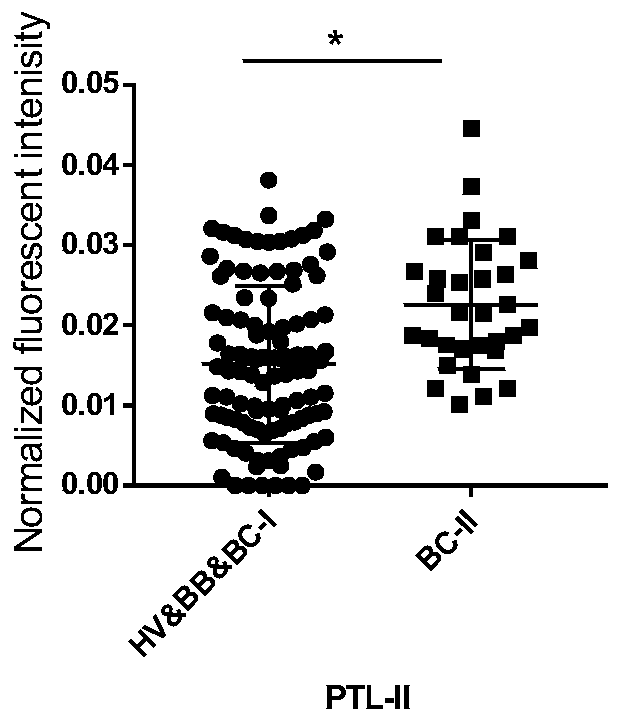
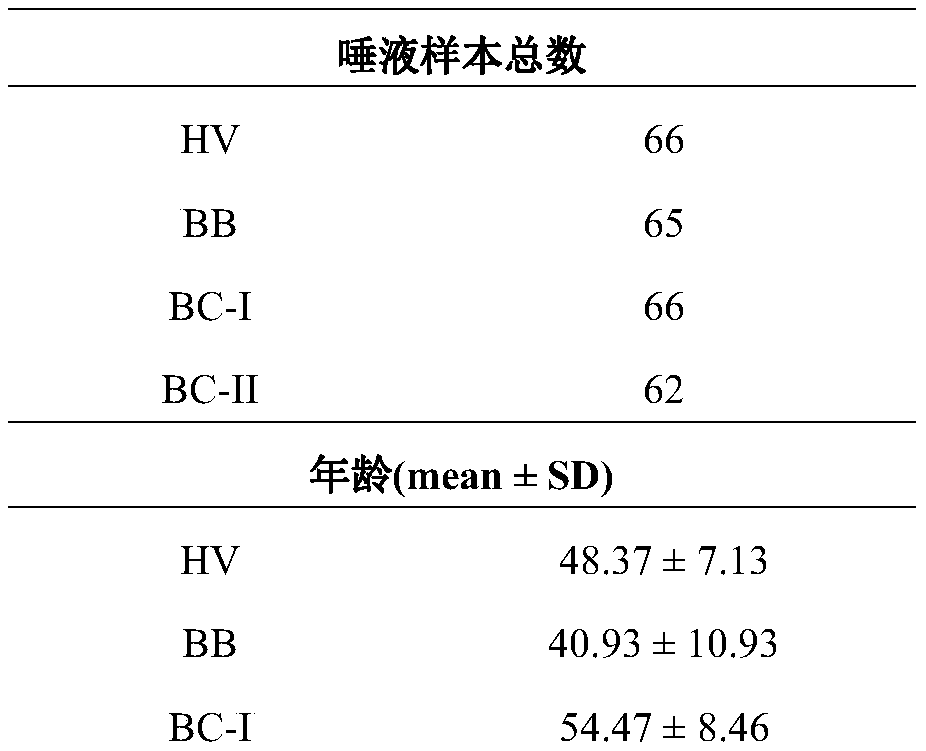
[0054] Previous studies have found that there are significant differences between breast cancer stage II salivary glycoprotein glycoforms and healthy volunteers, and the sugar chains recognized by PTL-II are significantly different between HV&BB&BC-I and BC-II groups ( figure 1 ). Based on this, we designed and completed the following experiments.
[0055] The relevant verification experiments and analysis of the present application are introduced in detail below, and the inventor's specific research and development process is not limited thereto.
[0056] 1. Research methods:
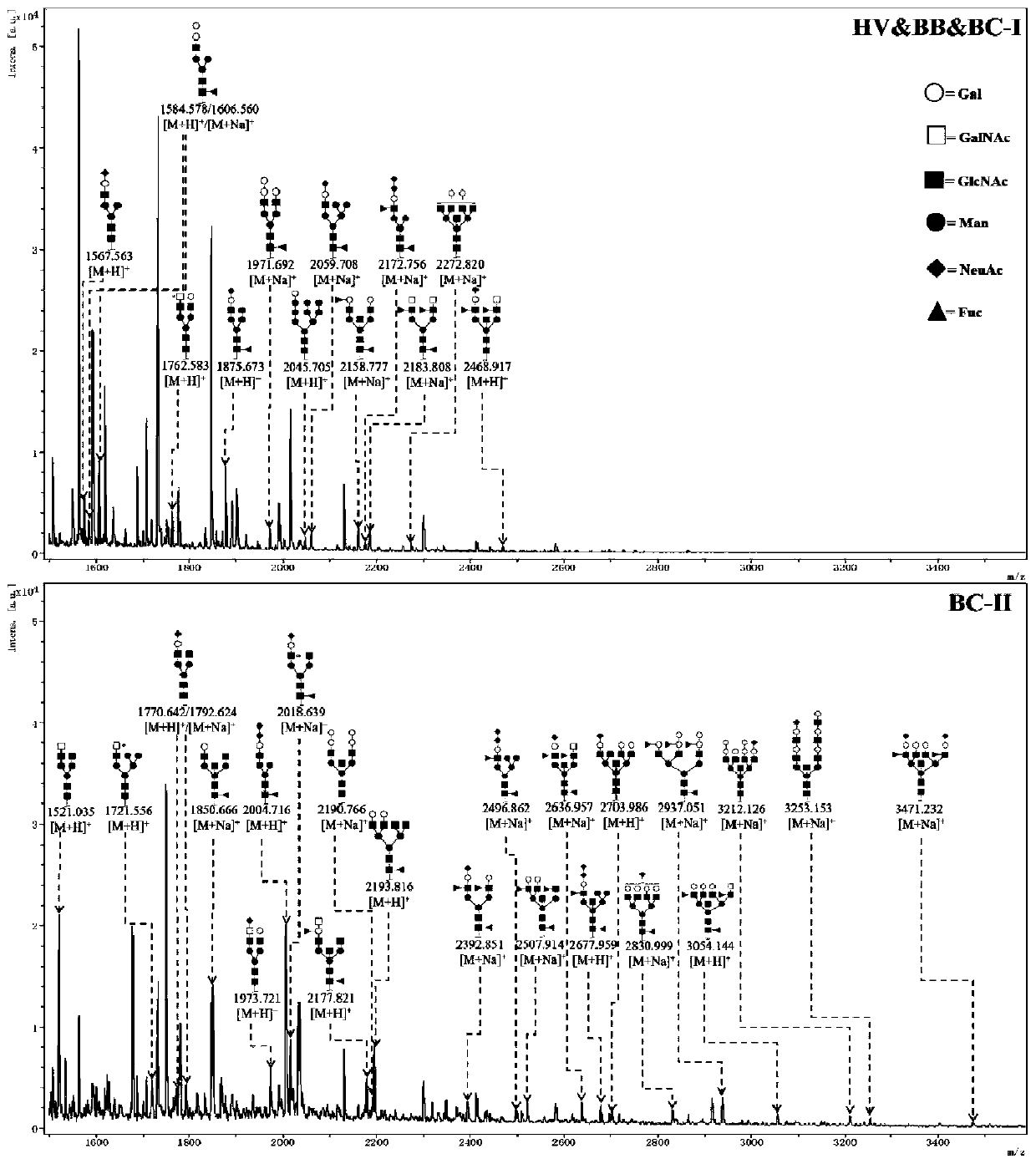
[0057] A lectin magnetic particle complex was prepared by coupling lectin PTL-II to the surface of iron ferric oxide nano-magnetic particles to enrich the specific glycoproteins in the mixed samples of saliva from each group, and the specific sugars were separated by PNGase F enzyme digestion method N-sugar chains on proteins, and mass spectrometric identification of sugar chains by Matrix-Assisted L...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com