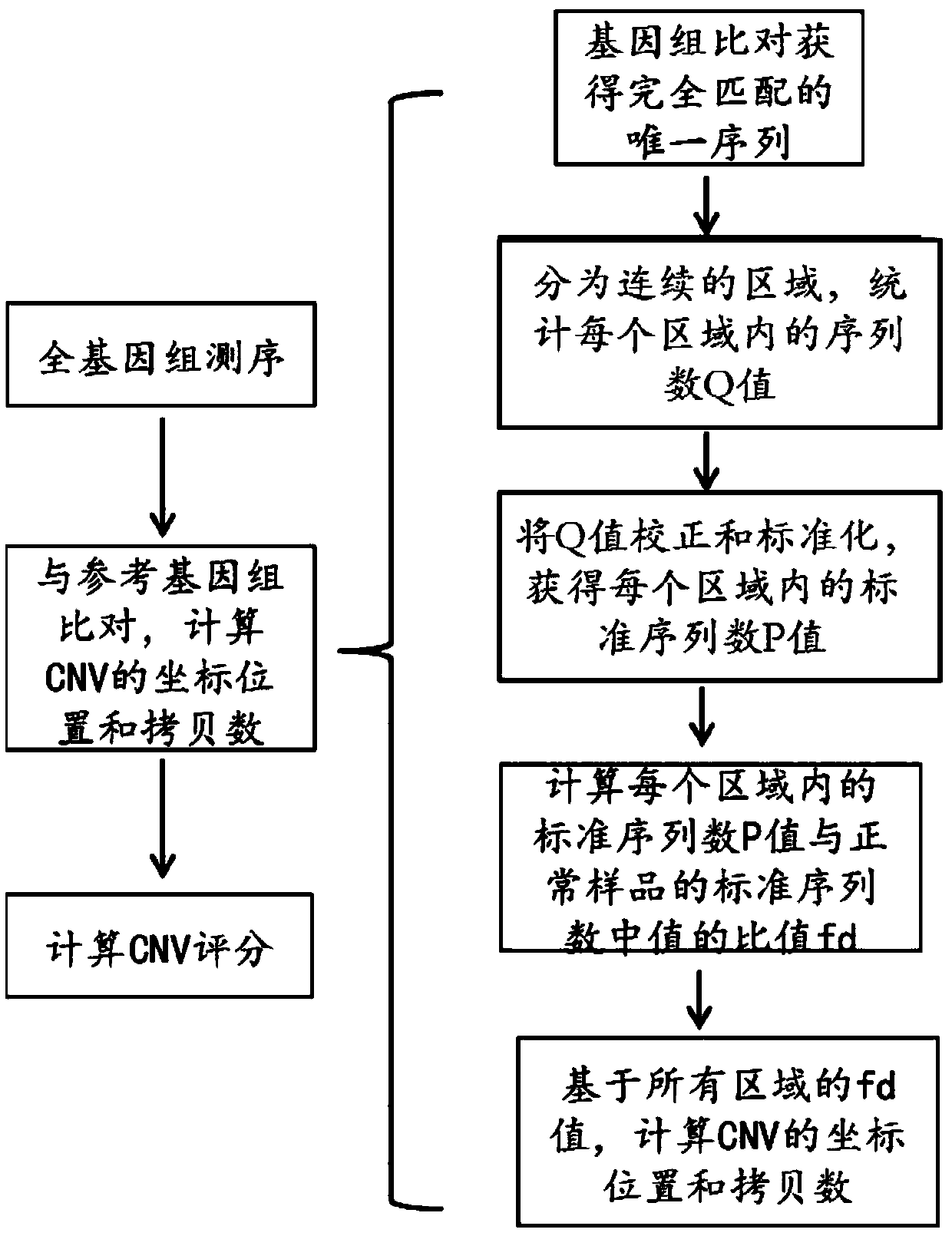
Detection method of whole genome copy number variation and application of the method
A copy number variation and genome-wide technology, applied in the field of cancer diagnosis devices, can solve the problems of inability to fully reflect the degree of genome disorder, difficulty in effectively diagnosing tumors, and single parameters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1 Determination of the genome-wide CNV score of the sample according to the detection method of the present invention
[0075] 1. Sample collection
[0076] Between January 2017 and March 2018, tumor tissue samples were obtained from patients through surgery at Beijing Hospital and the Affiliated Hospital of Zhengzhou University. The patients were pathologically diagnosed lung cancer patients and were not treated for lung cancer. Specifically, the patient had the following lung cancers: lung cancer in situ (AIS), minimally invasive lung cancer (MIA), invasive lung cancer (IA), malignant pleural effusion (MPE) and metastatic nodule (MN). Among them, AIS and MIA belong to low-invasive lung cancer, and IA belongs to high-invasive lung cancer. A total of 71 patients' tumor tissue samples and 15 patients' paracancerous samples (ie, non-cancerous samples) were obtained. Tumor tissue samples and paracancerous samples obtained from patients through surgery were fixed...
Embodiment 2
[0093] Example 2: Diagnosis of lung cancer, its invasiveness and cancer grade based on the whole genome CNV detection method of the present invention
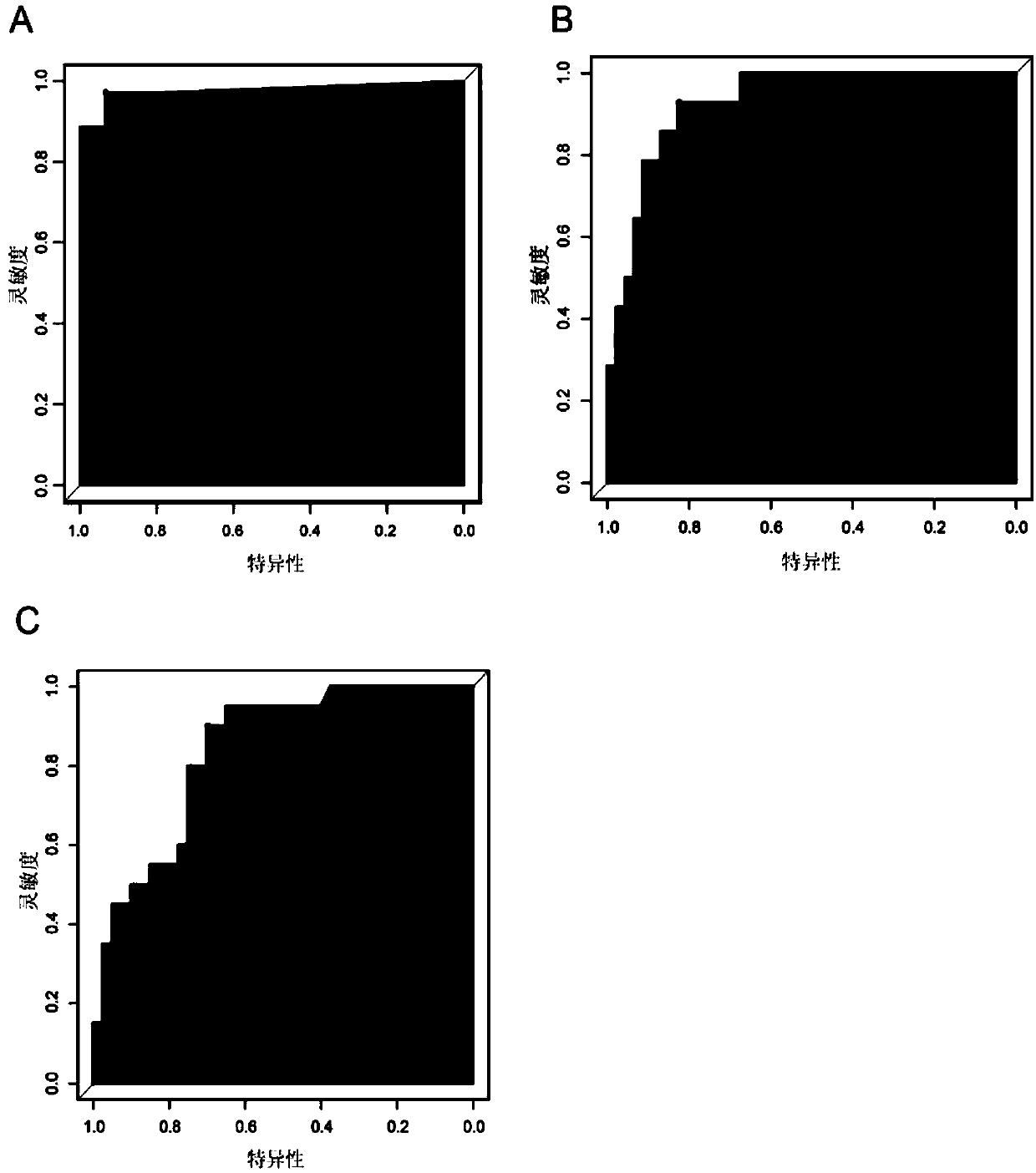
[0094] 1. draw the ROC curve according to the CNV score of 71 tumor tissue samples obtained in embodiment 1 and 15 paracancerous samples, to determine the threshold (see figure 2 A-2B). Such as figure 2 As shown in A, when the threshold of CNV score was set at 0.015, it had the highest accuracy in distinguishing cancer samples from normal samples, with a sensitivity and specificity of 93.3% and 97.2%, respectively. Such as figure 2 As shown in B, when the threshold of CNV score was set at 1.940, it had the highest accuracy in distinguishing low-invasive cancer samples from high-invasive cancer samples, with a sensitivity and specificity of 82.6% and 92.9%, respectively. Such as figure 2 As shown in C, when the threshold of CNV score is set at 2.550, it has the highest accuracy in distinguishing low-grade (grade I, II) a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com