Neospora caninum specific PCR detection kit and preparation method and application
A technology of Neospora caninum and detection kit, which is applied in biochemical equipment and methods, microbe determination/inspection, DNA/RNA fragments, etc. It can solve the problems that the detection of the disease cannot meet the actual needs and the quarantine diagnosis lags behind.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
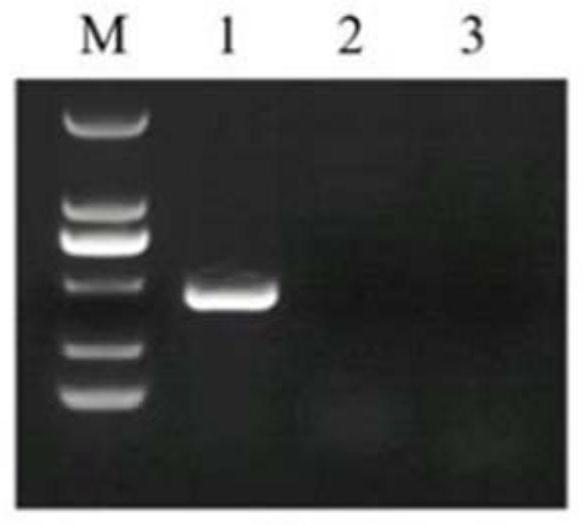
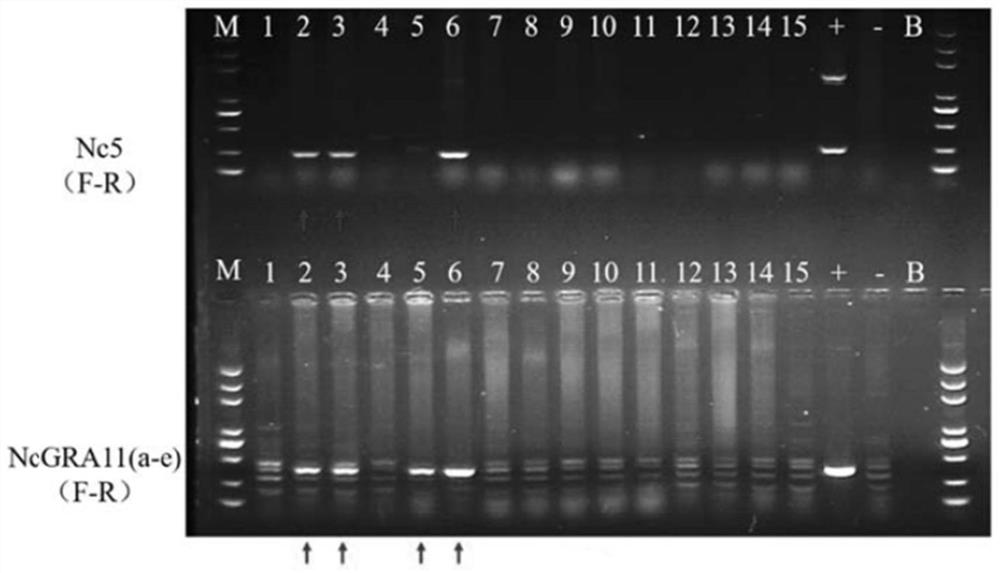
[0027] According to the principle of PCR amplification, the present invention designs and synthesizes a group of PCR primers for amplifying the specific gene sequence SEQID NO.1 of Neospora caninum. The synthesis of the primers is entrusted to Beijing Ruiboxing Biotechnology Co., Ltd. After primer synthesis, use sterilized distilled water to make 100mM stock solution (stock solution), and then use ddH 2 O was diluted to 10 mM as a working solution. The primer sequences are as follows:
[0028] Upstream primer F: 5'-CGCACAAGAAACCGAGGAA-3' (SEQ ID NO.2)
[0029] Downstream primer R: 5'-GAAGGCGAGAAGCCCAAGT-3' (SEQ ID NO.3)
Embodiment 2
[0031] kit assembly
[0032] 1) 10×PCR reaction solution: 20 μL of 300 mM dNTPs (purchased from TAKARA, Japan), 2.42 mg of Tris (purchased from Pragma, USA), 7.45 mg of KCl (purchased from Sigma, USA), MgCl2 (purchased from Sigma, USA) 0.258mg, 200U Taq enzyme (purchased from Japan TAKARA company) 20μL, add 150μL ddH 2 O, adjust the pH to 8.5 with HCl, use ddH 2 O to dilute to 200 μL.
[0033]2) Preparation of positive control DNA: the positive control DNA is Neospora caninum genomic DNA, and the cultured Neospora caninum is counted with a cell counting plate and counted with 10 7 Genomic DNA was extracted from each Neospora caninum and diluted to 100 μL with water.
[0034] 3) Assembly of the kit:
[0035] Assemble the kit as follows (the entire operation requires a sterile environment):
[0036] 10×PCR reaction solution 200μL
[0037] 10mM upstream primer F 50μL
[0038] 10mM downstream primer R 50μL
[0039] Positive control DNA 100μL
[0040] wxya 2 O 2mL
Embodiment 3
[0042] The using method of kit of the present invention is as follows:
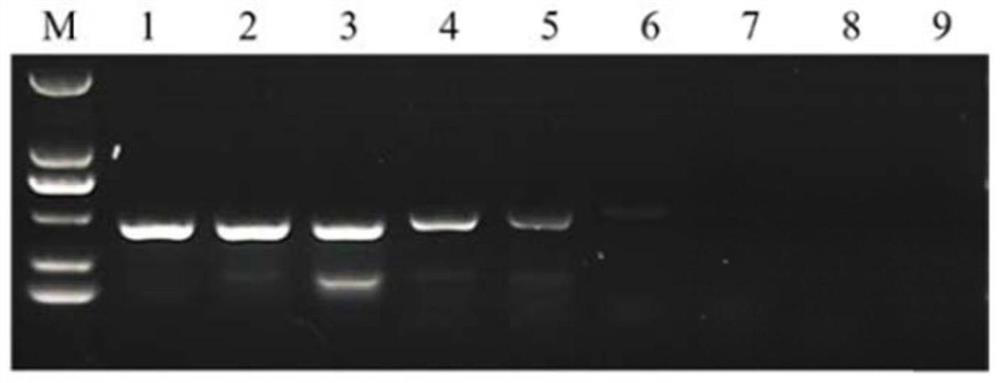
[0043] Take the sample to be tested, extract genomic DNA according to conventional methods, and perform PCR amplification with the reagents in this kit. The PCR reaction sample loading system is as follows, the total reaction system is 20 μL (the amount of sample DNA is determined according to the concentration of the extracted genomic DNA, and finally with ddH 2 O to make up 20 μL; positive control volume is 1 μL):
[0044] 10×PCR reaction solution: 2μL
[0045] Determine the sample DNA according to the amount of extracted DNA
[0046] Upstream primer F (10mM): 1μL
[0047] Downstream primer R (10mM): 1μL
[0048] wxya 2 O: Make up to 20 μL
[0049] Add the negative and positive controls to the marked tubes respectively, then add the samples one by one and mark them, and place them in the PCR machine. The PCR reaction conditions were: pre-denaturation at 94°C for 4 min, denaturation at 94°C for 30...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com