Transcription factor binding site prediction method based on depth convolution automatic encoder
An autoencoder and binding site technology, applied in the field of computer technology and bioinformatics, can solve the problems of insufficient model generalization ability, limited prediction level of different transcription factors, affecting the performance of prediction models in TFBS, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0032] In order to facilitate those skilled in the art to understand the technical content of the present invention, the content of the present invention will be further explained below in conjunction with the accompanying drawings.
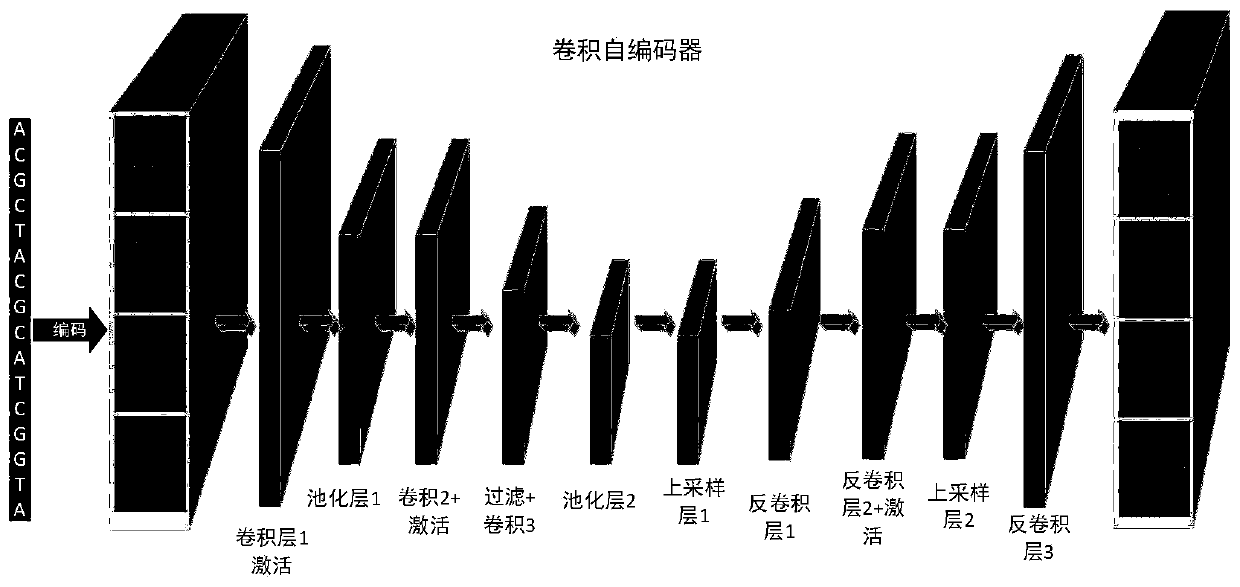
[0033] In the present invention, considering the spatial and sequential features of DNA sequences, we design a hybrid deep neural network that integrates a convolutional autoencoder and a high-speed fully-connected MLP at this stage. Convolutional Neural Networks (CNNs) are specialized versions of Artificial Neural Networks (ANNs) that employ a weight-sharing strategy to capture local patterns in data such as DNA sequences. Including the preliminary prediction algorithm, feature extraction and model establishment of transcription factor binding sites for the preprocessed DNA sequence data, the overall flow chart of the system is as follows figure 1 shown. The following will introduce each in detail:
[0034] The invention aims to make full use ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com