Rapid division method for big data gene sequencing files
A gene sequencing and big data technology, applied in the field of high-performance computing, can solve problems such as affecting bwa results and inconsistency in comparison results, and achieve the effects of improving the division speed, reducing the number of reading and writing, and eliminating comparison errors.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
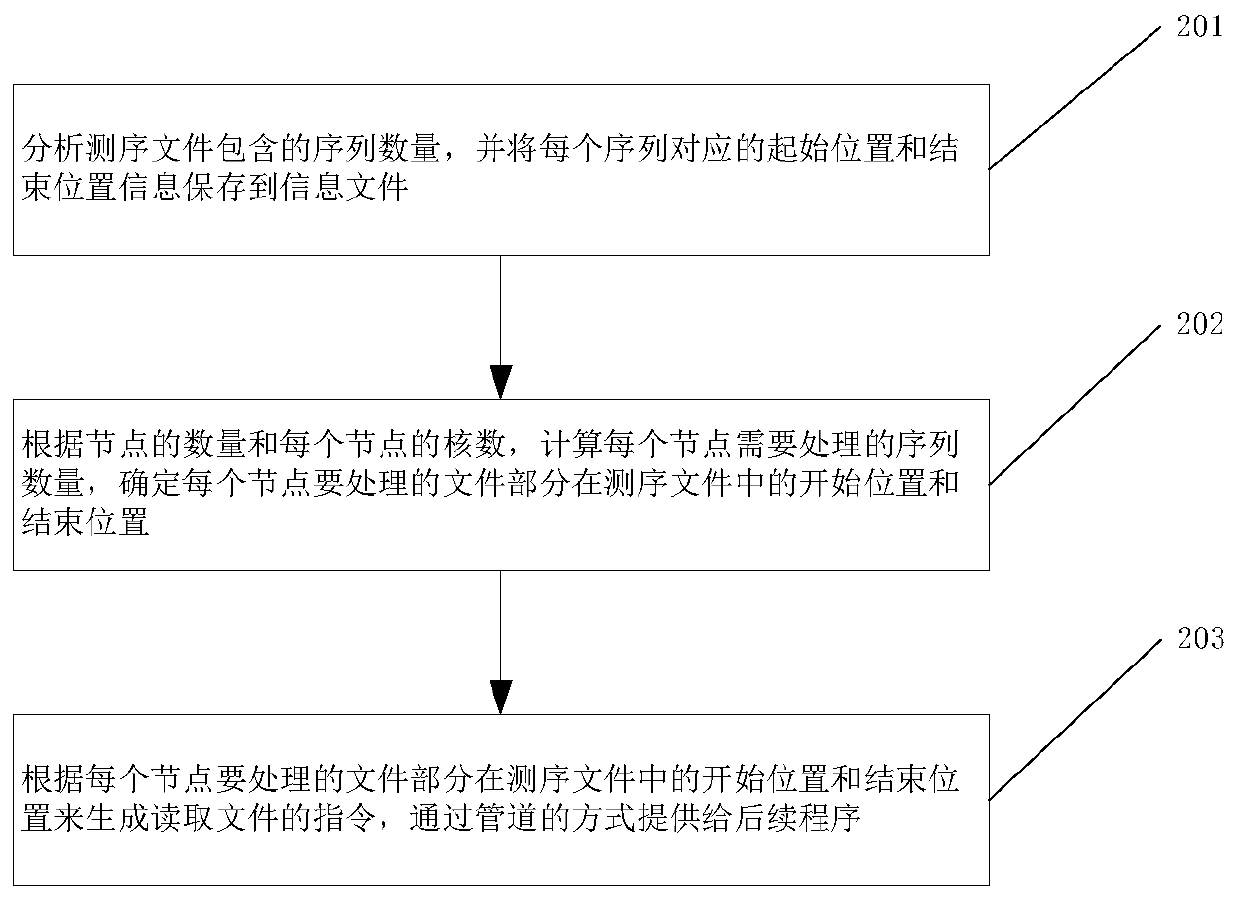
[0041] Before describing this method in detail, the format of the fastq file is briefly introduced. The fastq file is a text file with a sequence every four lines. The first line is the name information of the sequence, the second line is the base sequence, the third line is the description information, and the fourth line is the quality score information of the sequence. Each sequence is not exactly the same length. Sequencing files are divided into single-end sequencing files and paired-end sequencing files. A single-end sequencing file contains only one file, and a paired-end sequencing file contains a pair of files, and each sequence in this pair of files corresponds to each other.
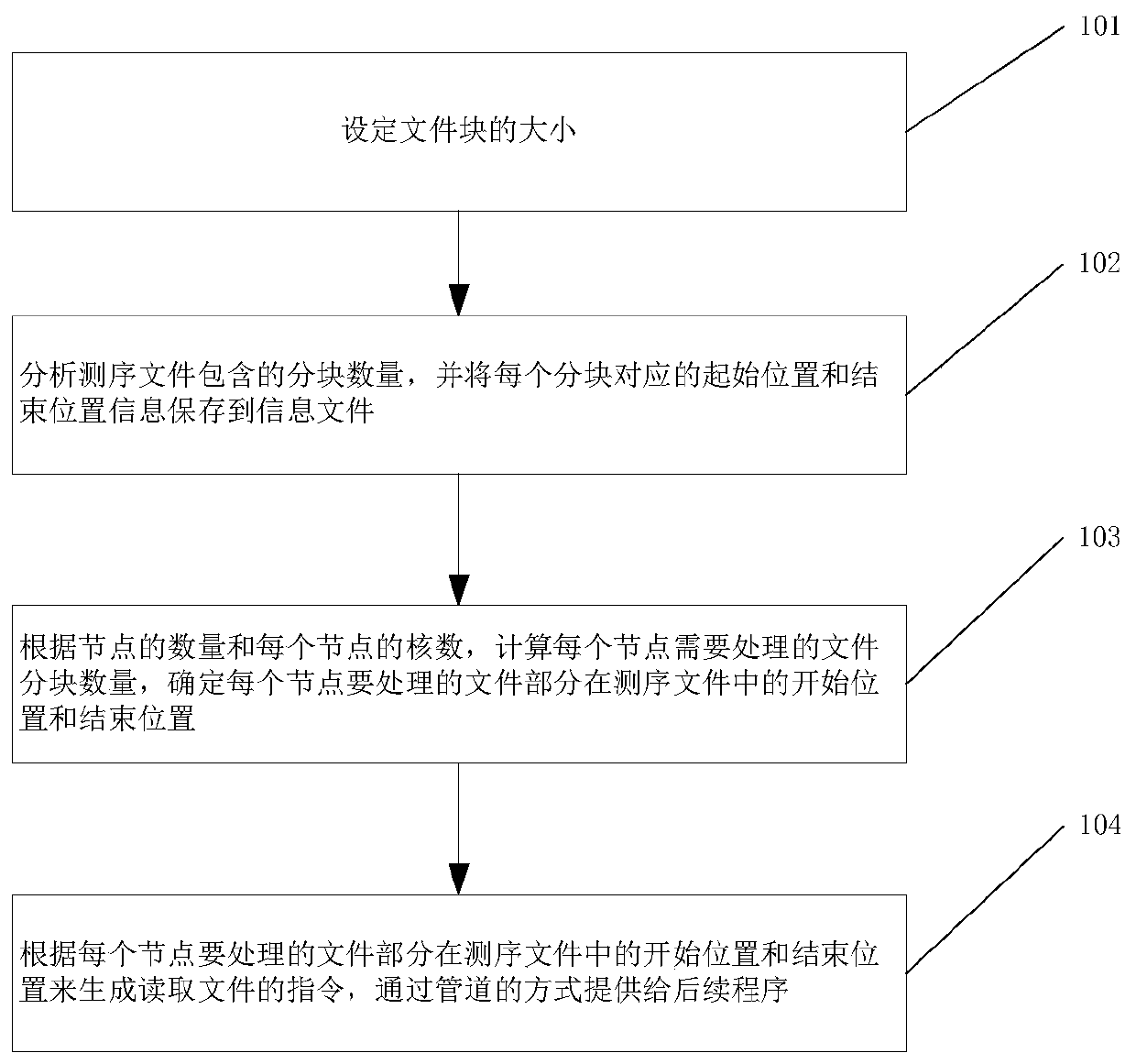
[0042] According to an embodiment of the present invention, combining figure 1 Introduce the method of partition by block, which includes the following steps.
[0043] Step 101: Set the size of the file block, preferably, the value range can be between 1M and 100M.
[0044] The inventor's r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com