Nucleic acid composition for detecting novel coronavirus COVID-19 and application
A technology for COVID-19 and nucleic acid composition, applied in the field of nucleic acid composition for the detection of novel coronavirus COVID-19, can solve the problems of low sensitivity, complicated operation, long diagnosis time, etc. fast effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] This embodiment provides a method for detecting the N gene of novel coronavirus COVID-19, comprising the following steps:
[0064] First, in order to detect the expression of the new coronavirus RNA, according to the nucleotide sequence of the N gene of the new coronavirus COVID-19 retrieved from the NCBI and Genbank databases, a conservative sequence of the N gene of the new coronavirus COVID-19 was chemically synthesized, and transfected Infect the cells, artificially prepare the pseudovirus, extract and serially dilute, and the diluted RNA is used as a template for transcription-mediated amplification (TMA).
[0065] Second, design and synthesize TMA upstream and downstream primers for amplifying the N gene of COVID-19 by selecting a highly conserved segment without secondary structure. Wherein, the sequence of the upstream primer N-t7F is: 5'-AATTTAATACGACTCACTATAGGGtgtaggtcaaccacgttccc-3'. Among them, capital letters are T7 promoter sequences, and lowercase letter...
Embodiment 2
[0073] The present embodiment provides a method for rapid detection and detection of the Orf1ab gene of novel coronavirus COVID-19, comprising the following steps:
[0074] First, pseudoviruses are artificially prepared as templates for transcription-mediated amplification (TMA).
[0075]Second, design and synthesize upstream and downstream primers for TMA amplification of the Orf1ab gene of COVID-19. Wherein, the upstream primer Orf1ab-t7F is: 5'-AATTTAATACGACTCACTATAGGGgcattctgtgaattataagg-3'. Among them, the 24 bases at the 5' end of Orf1ab-t7F are the added T7 phage promoter sequence, and the 20 bases at the 3' end are the specific sequence that hybridizes with the 5' end of the template RNA (+). The downstream primer Orf1ab-R is: 5'-atactgctgccgtgaacatg-3', which is a sequence hybridizing with RNA (-).
[0076] Third, by designing and synthesizing primers, using the method of annealing the primers in boiling water and in vitro transcription, a sgRNA targeting the negati...
Embodiment 3
[0083] This embodiment provides a method for detecting the Orf1ab gene of the new coronavirus COVID-19 with TMA and Cas13a "two-in-one", including the following steps:
[0084] First, the templates, primers and nucleic acid probes required in Example 3 can be prepared according to Example 2.
[0085] Second, establish a "two-in-one" reaction system (50 μL) of TMA combined with Cas13a. The reagents and specific concentrations involved in this reaction are: RNA (+) fragment to be detected, TMA upstream and downstream primers Orf1ab-t7F (1μM) and Orf1ab-R (1μM), 2000-4000U M-MLV reverse transcriptase, 500-2000U T7 RNA polymerase, 4U RNase inhibitor, 2μM cas13a protease, 400nM Orf1ab-sgRNA, 0.5μM OligoRNA-Cas13a-probe, TC reaction buffer (20-50mM Tris-HClpH7.5@25℃, 20-40mM KCl, 1- 5mM MgCl 2 , 1-5mM rNTPs, 1-5mM dNTPs, 20-50% glycerol, 0.5-1mMDTT).
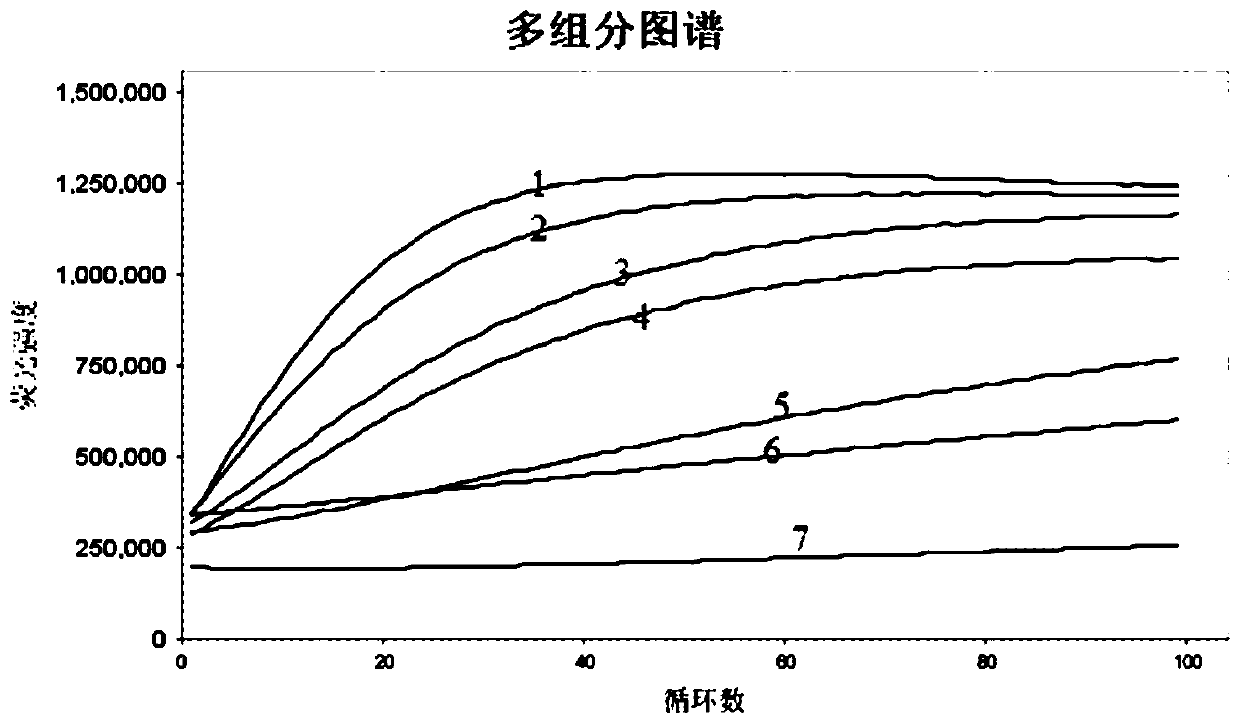
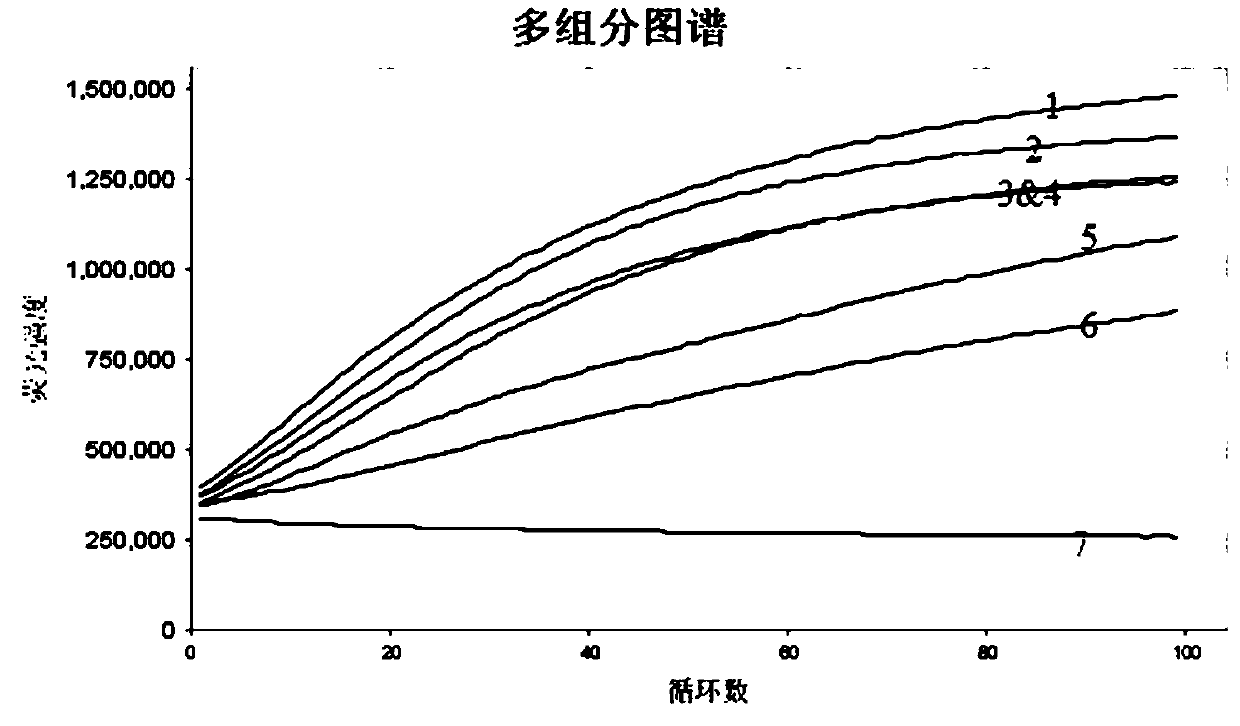
[0086] Thirdly, after the components of the reaction system were gently vortexed and mixed, the reaction tube was reacted by a fl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com