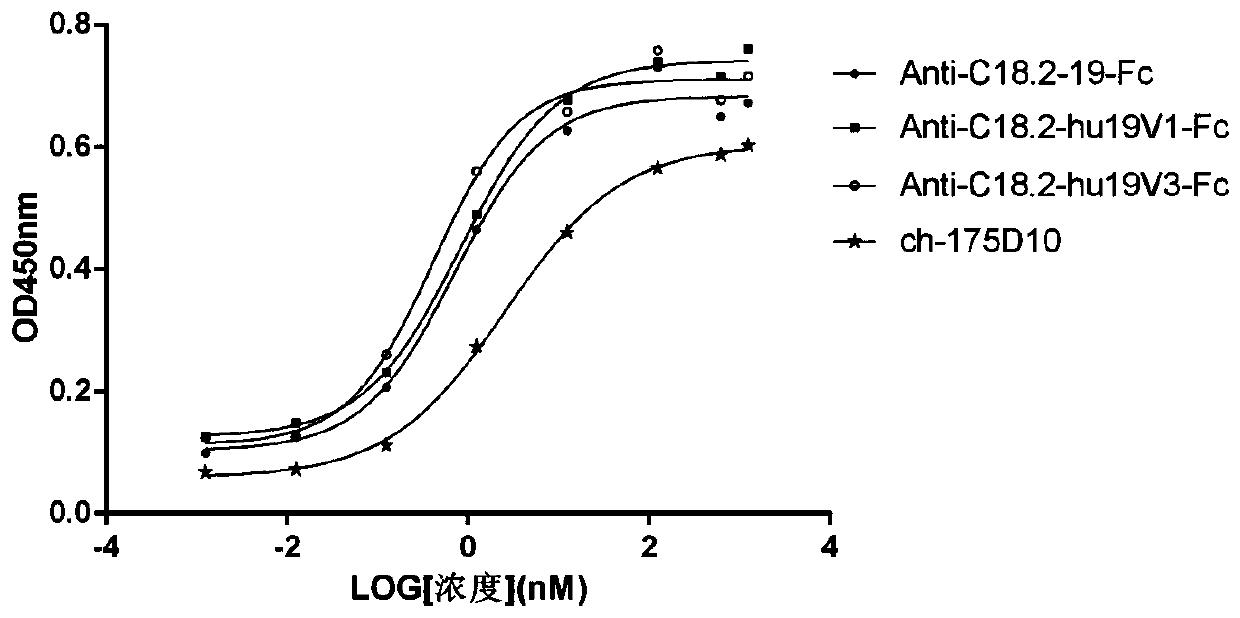
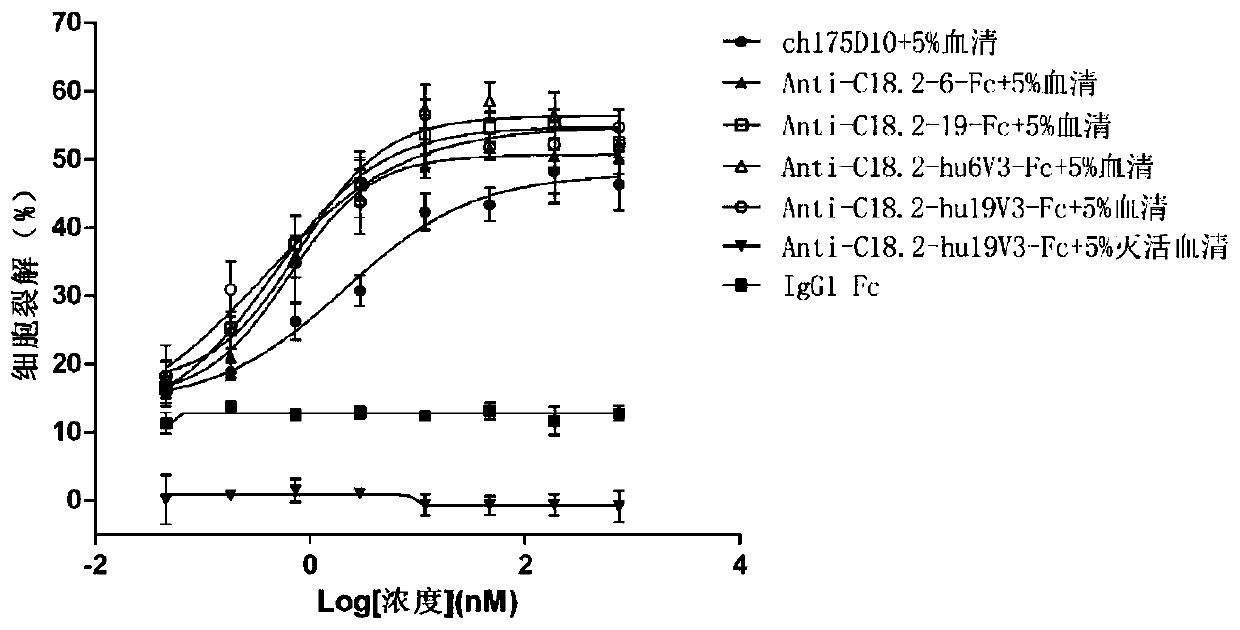
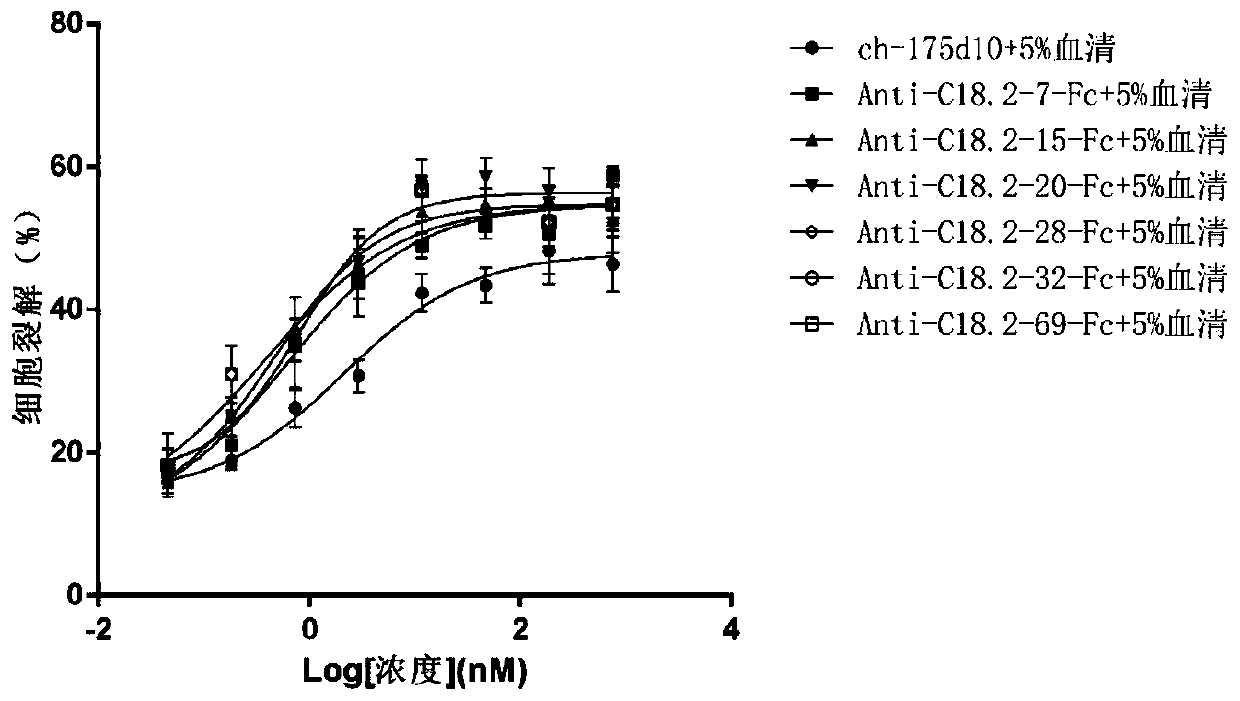
Anti-CLD18A2 nanobody and application thereof
A nanobody, FR1 technology, applied in the direction of antibody medical components, recombinant DNA technology, antibody mimics/scaffolds, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0143] Example 1: Construction and detection of cell lines expressing CLD18A2
[0144] The pCDNA3.1 vectors (Life Technologies) containing the full-length genes of CLD18A1 (amino acid sequence, SEQ ID NO.96) and CLD18A2 (amino acid sequence, SEQ ID NO.97), respectively, were extracted with a plasmid extraction kit (Biomiga), The expression plasmid was sterile-filtered and electrotransfected into CHO-S cells, and G418 (sigma) was added for 96-well plating to construct CHO-S-CLD18A1 and CHO-S-CLD18A2 stably transfected cell lines, respectively. Pick the stable cell lines on the 96-well plate and gradually amplify them under the culture condition of adding G418, and use the Anti-Claudin18 antibody [34H14L15] (abcam) to detect the positive expression cell line CLD18A1 by dot blotting. and CLD18A2. The same method was used to construct NUGC-4-CLD18A1 and NUGC-4-CLD18A2. Gastric adenocarcinoma NUGC-4 was purchased from Wuhan Jinkairui Co., Ltd. The gastric adenocarcinoma NUGC-4 did...
Embodiment 2
[0145] Example 2: Anti-CLD18A2 nanobody library construction
[0146] The CHO-S-CLD18A2 in Example 1 was stably transfected with 1.0×10 7 A healthy alpaca (Vicugna pacos) was immunized with cells per ml, and immunized with 1ml of Freund's complete adjuvant (sigma), and then immunized again after 21 days. A total of 3 times of immunization stimulated B cells to express antigen-specific nano Antibody. One week after the third immunization, 30ml of alpaca blood was collected with a vacuum blood collection tube, lymphocytes were separated by lymphocyte separation medium (Tianjin Haoyang Huake Biotechnology Co., Ltd.), and total RNA was extracted by Trizol method. 3 μg of total RNA was reverse-transcribed into cDNA using a reverse transcription kit (Invitrogen) according to the instructions, and VHH was amplified using nested PCR and the following primers: the upstream primer of the first round of PCR 5'-CTTGGTGGTCCTGGCTGC-3' (SEQ ID NO.110) and downstream primer 5'-GGTACGTGCTGTT...
Embodiment 3
[0147] Example 3: Screening and Identification of Anti-CLD18A2 Nanobodies
[0148] 3.1 Screening of Anti-CLD18A2 Nanobodies:
[0149] The constructed Anti-CLD18A2 nanobody library was packaged with the helper phage M13KO7 (NEB), and the titer of the recombinant phage displayed in the display library was 5.7×10 13 PFU / ml. Take 18ml, 7×10 5 pcs / ml of CHO-S-CLD18A2 and 15ml, 3×10 6 CHO-S cells / ml, after centrifugation at 300g for 5 minutes at 4°C, remove the medium supernatant, resuspend the cells in PBS, centrifuge again and block with 2% skimmed milk powder (diluted in PBS) for 1 hour at room temperature. The recombinant phage library was approximately 5.7×10 11 Add PFU to the closed CHO-S cells (about 4.5×10 7 ), and incubated at room temperature for 30 minutes to perform two whole-cell subtractive screening methods. Add the supernatant to about 1.5 x 10 7 A well-blocked CHO-S-CLD18A2 stably transfected cells were incubated at room temperature for 1 hour for binding, an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com