Bombyx mori whole genome knockout vector library based on CRISPR/Cas9 system and construction method
A construction method and a knockout carrier technology, applied in the field of eukaryotic gene knockout
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0044] The Bombyx mori embryonic cell line (BmE) used in this example is a commonly used cell line in biological experiments (PMID: 17570024).
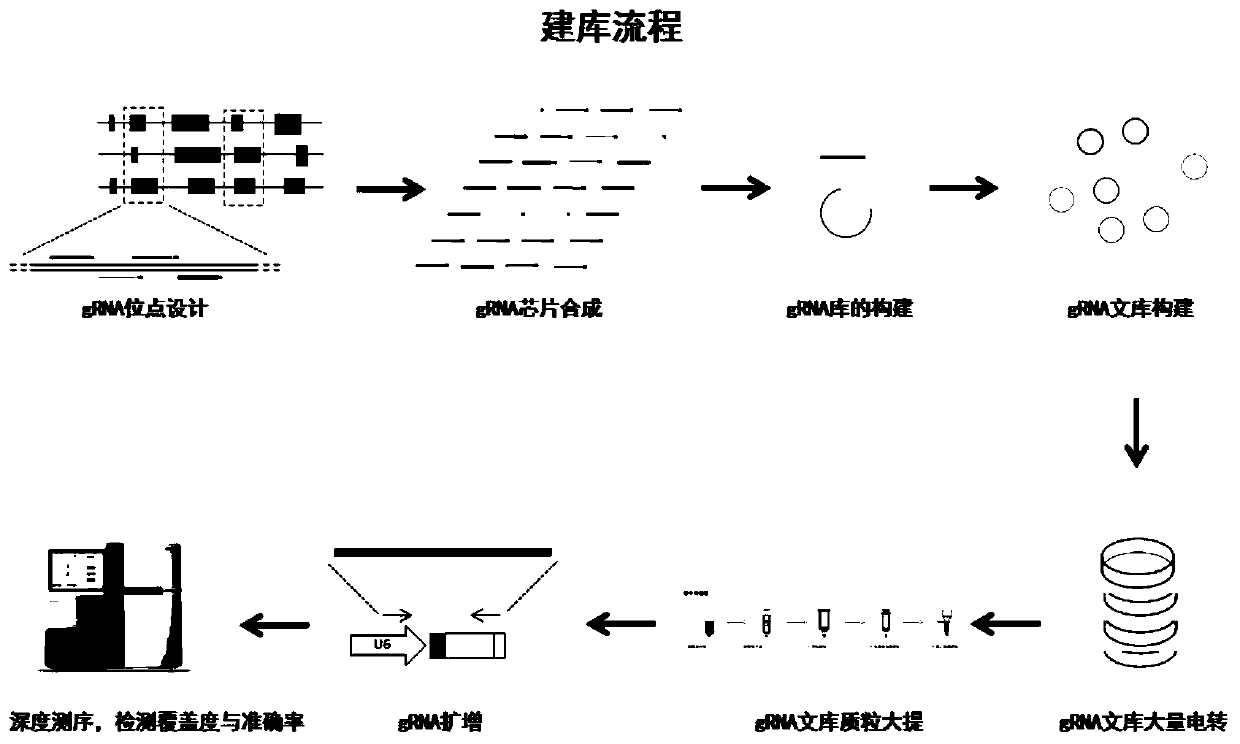
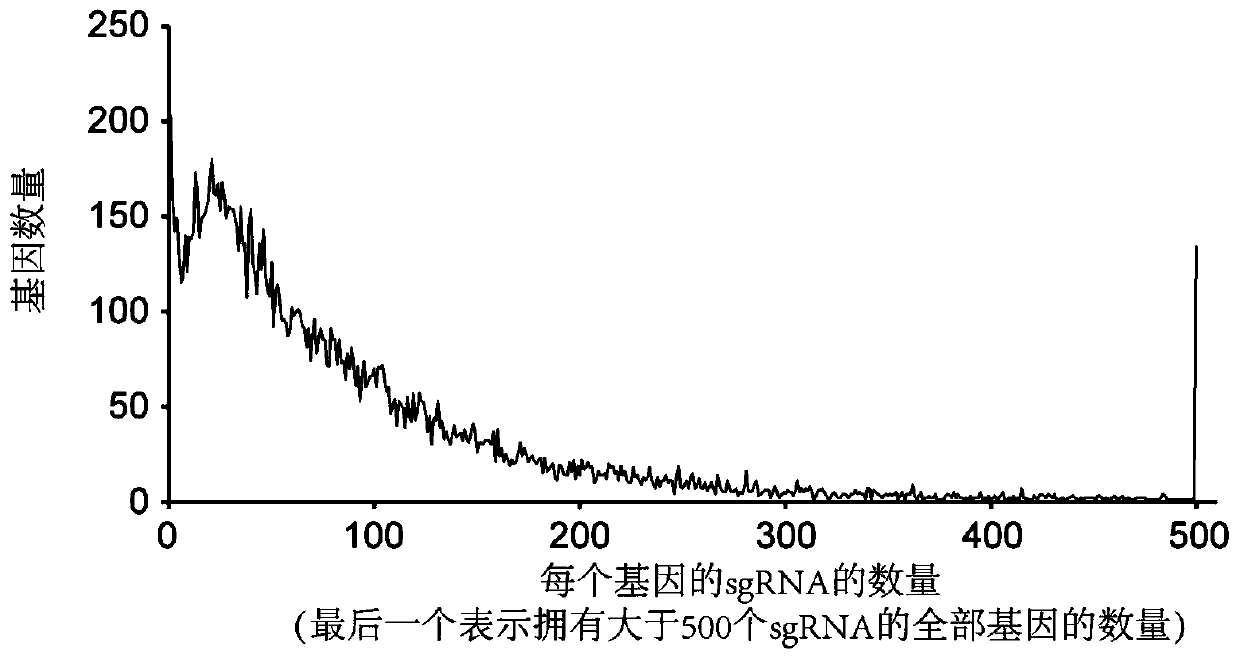
[0045] 1. The purpose of the present invention is to provide a vector library for silkworm genome-wide knockout based on the CRISPR / Cas9 system. The process of building a library is as follows: figure 2 shown.
[0046] 2. In order to realize the delivery of the silkworm CRISPR / Cas9 system, the present invention provides a delivery vector pB-CRISPR based on the piggyBac transposon system, its nucleotide sequence is shown in SEQ ID NO.1, and the vector map is shown in figure 1 . details as follows:
[0047] Using the piggyBac transposon system basic vector piggyBacModify (synthesized by Kingsray) as the initial vector, a piggyBac transposon system-mediated CRISPR / Cas9 gene knockout vector backbone was constructed, mainly including piggyBac transposable arms (including two piggyBac Transposon terminal inverted repeat sequence (invert...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com