Method and device for quantitatively detecting metagenome pathogens based on internal reference
A genome and pathogenic technology, applied in the field of pathogen detection, can solve the problems of not being able to fully demonstrate the multiple functions of internal reference quality control, unable to provide guidance, and not fully utilizing the role of internal reference quality control products in metagenomic sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
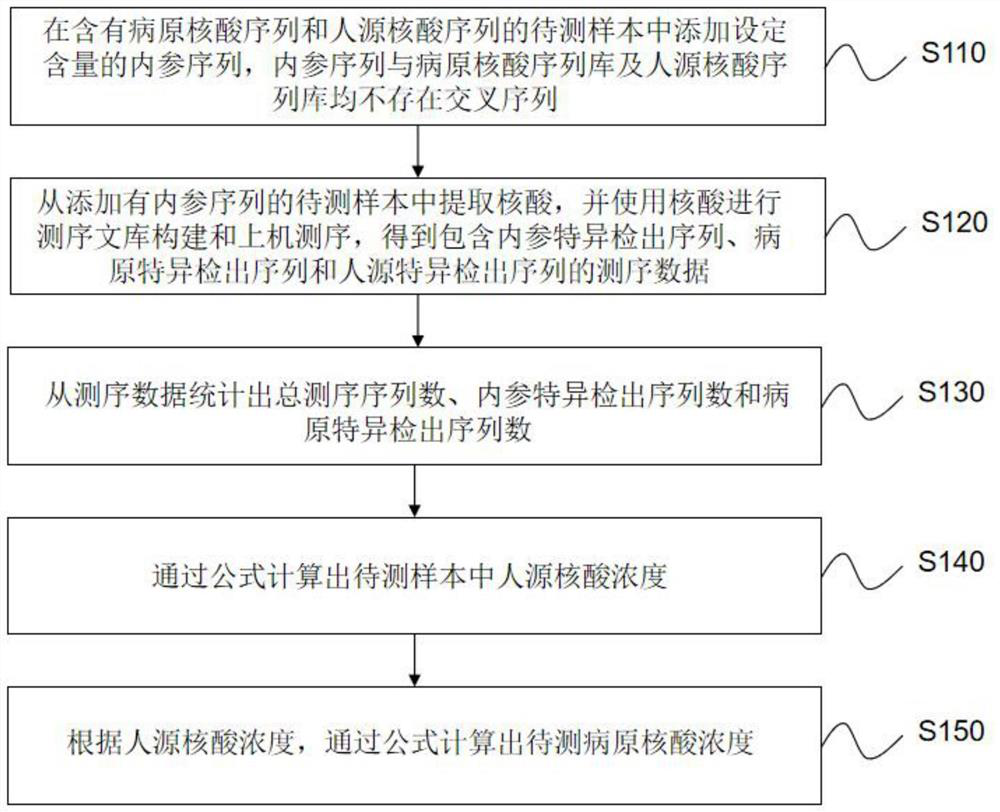
[0089] In this embodiment, the randomly generated specific sequence was used as the internal reference sequence for analysis.
[0090] This example is mainly used to demonstrate the accuracy of quantification of human nucleic acid content through internal reference sequences, and real clinical samples are used for evaluation. The main process is as follows:
[0091] 1. Internal reference screening
[0092] A random sequence generator was used to generate a simulated sequence to generate a random sequence of 300-500 bp, and the generated random sequence was analyzed by blast software for specificity.
[0093] First, the generated sequence is randomly cut into short sequences of 35 bp by information analysis software, and the short sequence is compared with the human sequence library and the pathogenic sequence library by blast software to screen for the most specific sequence, that is, no human sequence is compared. Sequences of the source sequence library and the pathogenic s...
Embodiment 2
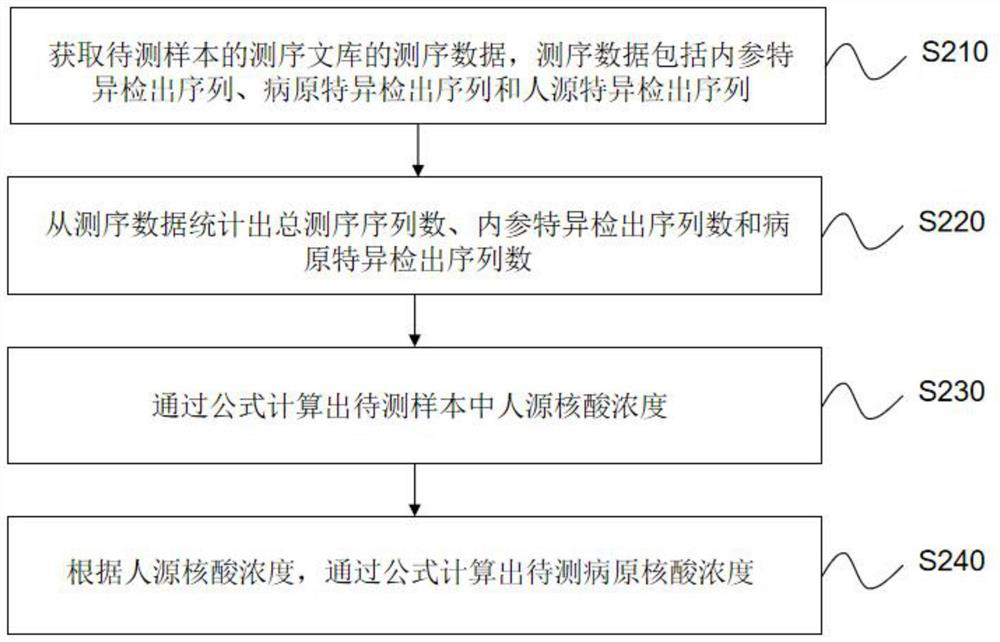
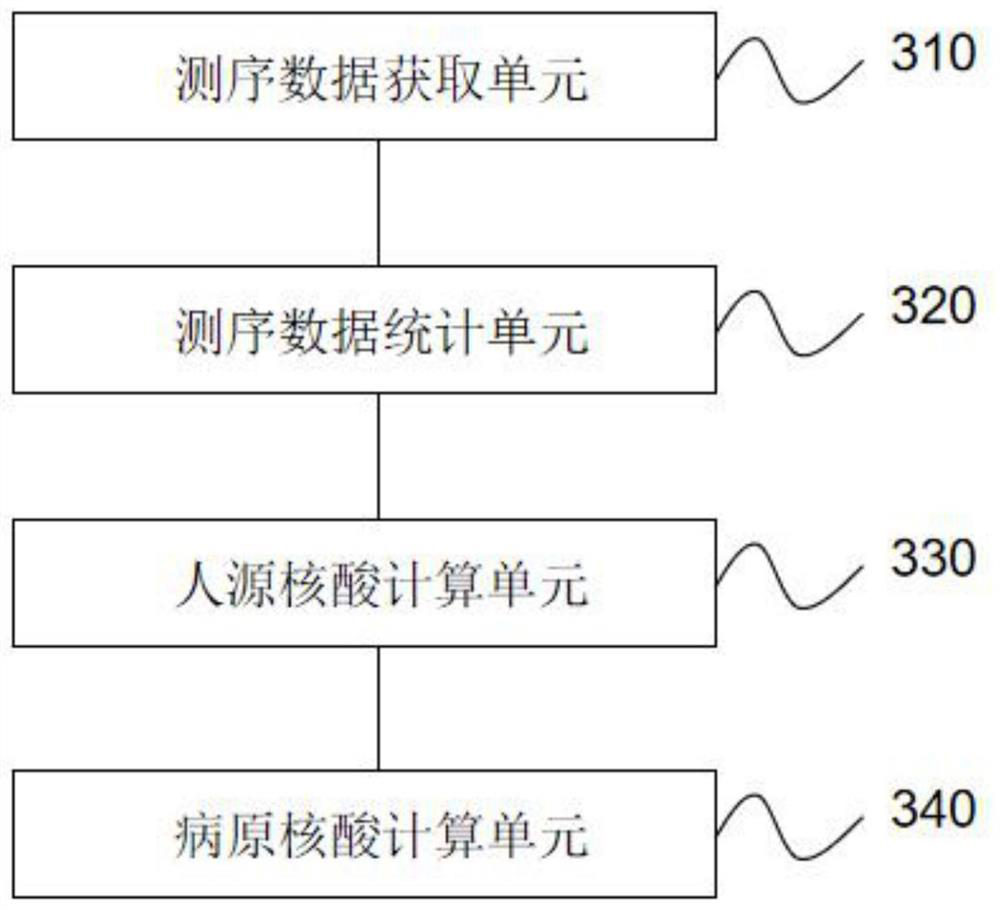
[0122] This example is mainly used to demonstrate the accuracy of the calculation of human nucleic acid content and pathogenic nucleic acid content through internal reference sequences, and simulated samples are used for evaluation. The main process is as follows:
[0123] 1. Select 30 cases of simulated cerebrospinal fluid samples, add known concentrations of human cells and different types of pathogens, add them according to the concentration of the internal reference sequence determined in Example 1, and perform nucleic acid extraction, library construction, computer sequencing, and data analysis Afterwards, the results are shown in Table 3 below:
[0124] table 3
[0125]
[0126]
[0127] 2. Carry out the calculation of human nucleic acid content according to the metagenomics theory derivation formula established by the present invention according to the detection value of the internal reference, taking the S21 sample as an example, the calculation method is as follow...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com