Fluorescence labeling-based uridine monophosphate acidification detection method
A technology of uridine monophosphate and fluorescent labeling, which is applied in the field of biochemistry, can solve the problems that have not received attention and the modification detection method has not been established, and achieve the effect of shortening the experimental time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0021] The present invention will be further described according to the following examples, and the mode of the present invention includes but not limited to the following examples.
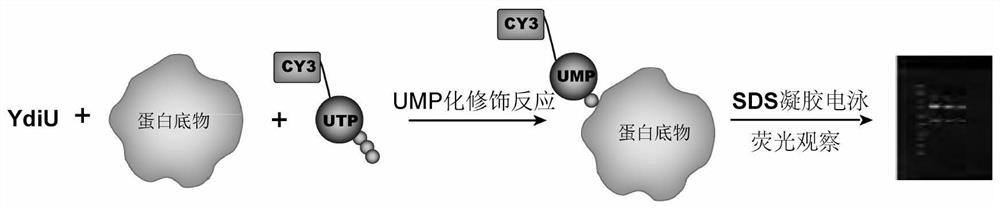
[0022] Such as figure 1 Shown, the present invention comprises the following steps:
[0023] A: Add YdiU protein and substrate to the reaction system, mix well, add 1 μl of CY3-X-UTP, and react for UMP modification at 37°C for 1 hour to obtain UMP-modified protein with CY3 label;
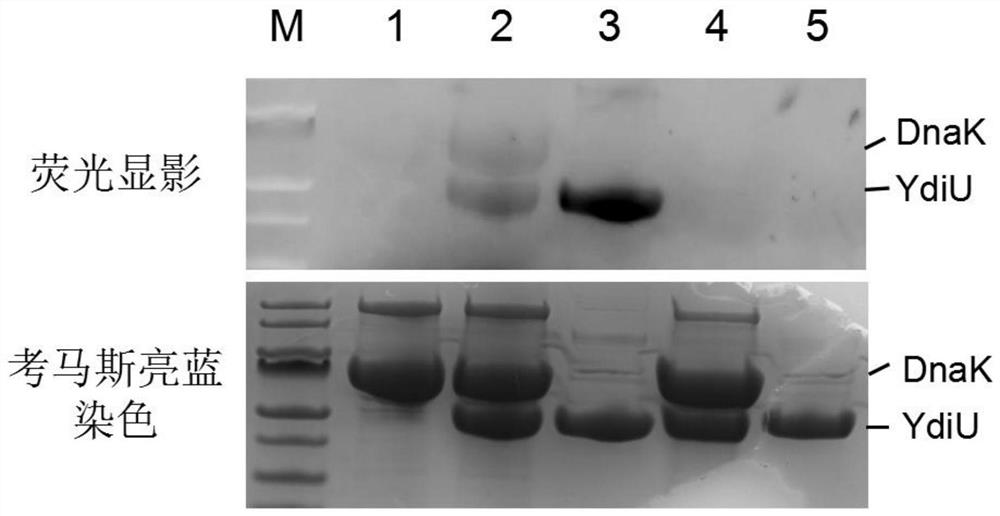
[0024] B. After adding the UMP-modified protein to the loading buffer, use 12% SDS-PAGE gel electrophoresis;
[0025] C Use a fluorescence imager to observe the gel under the emission of 565 nm after excitation at 553 nm.
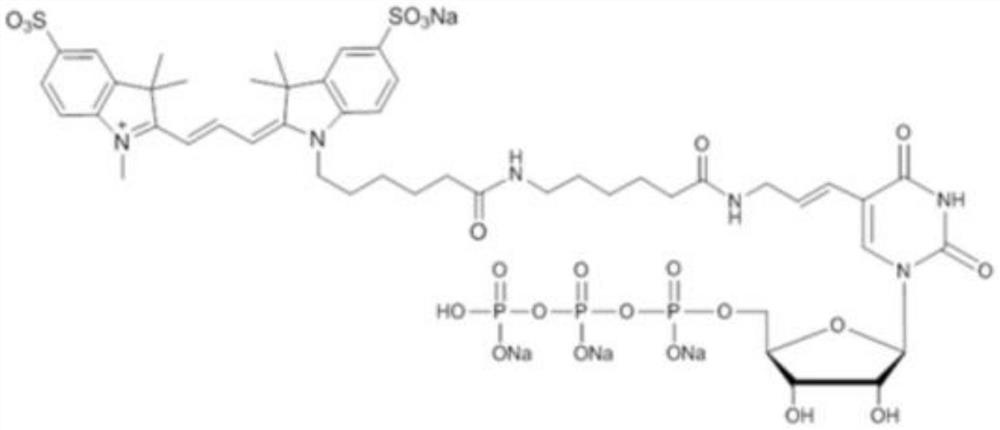
[0026] The molecular structure of the CY3-labeled UMP donor CY3-X-UTP used in the modification reaction is as follows:
[0027]
[0028] Based on this identification method, a kit is developed, which provides UMP transferase that catalyzes uridylation of monophosphate, fluorescently labeled UT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com