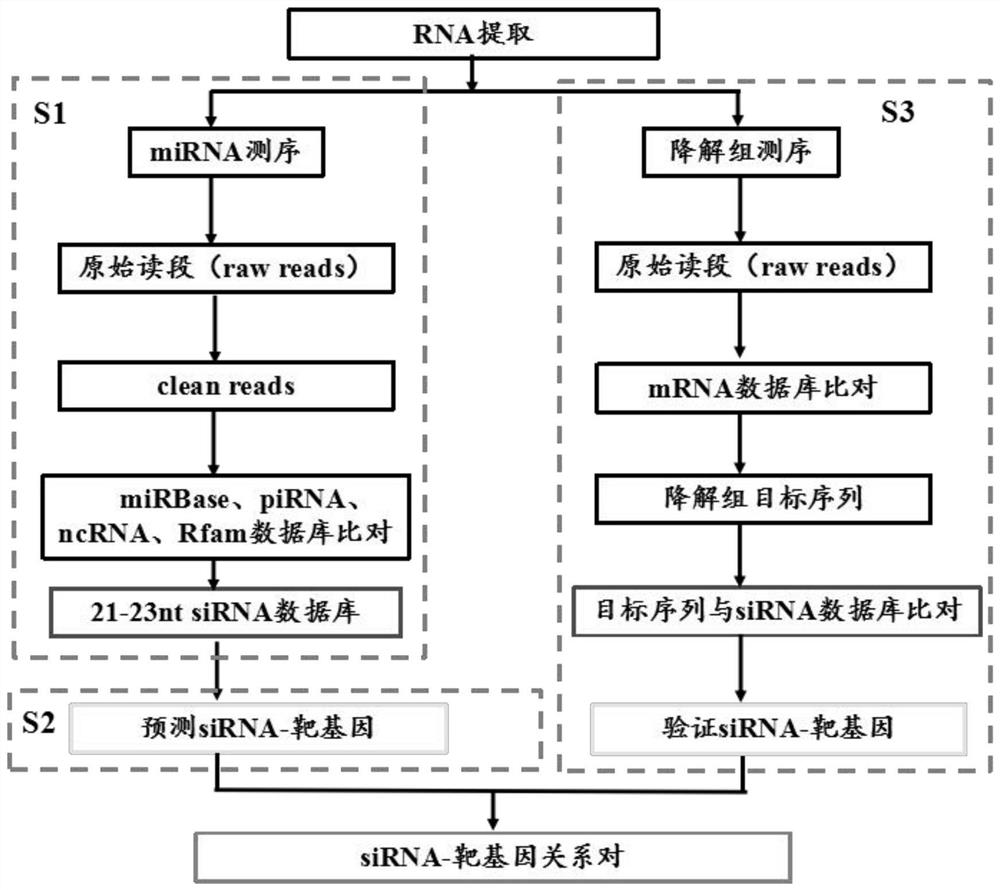
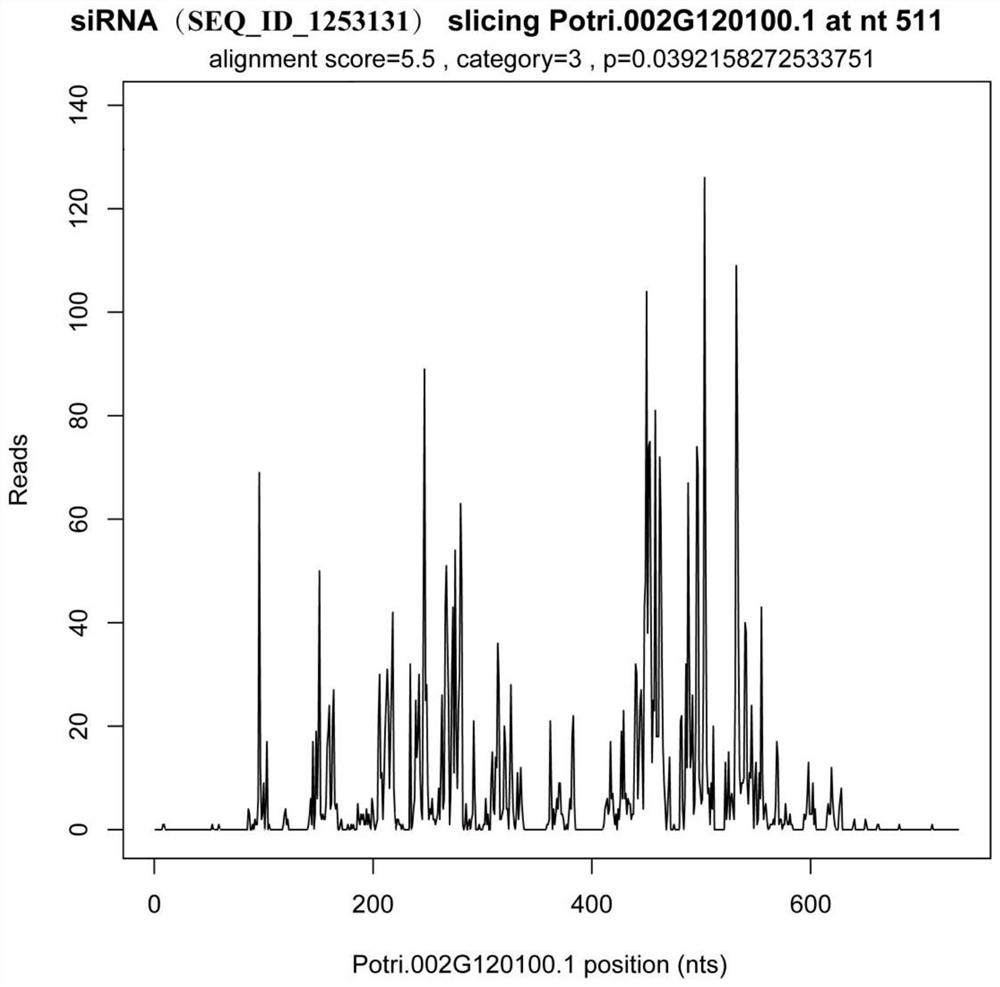
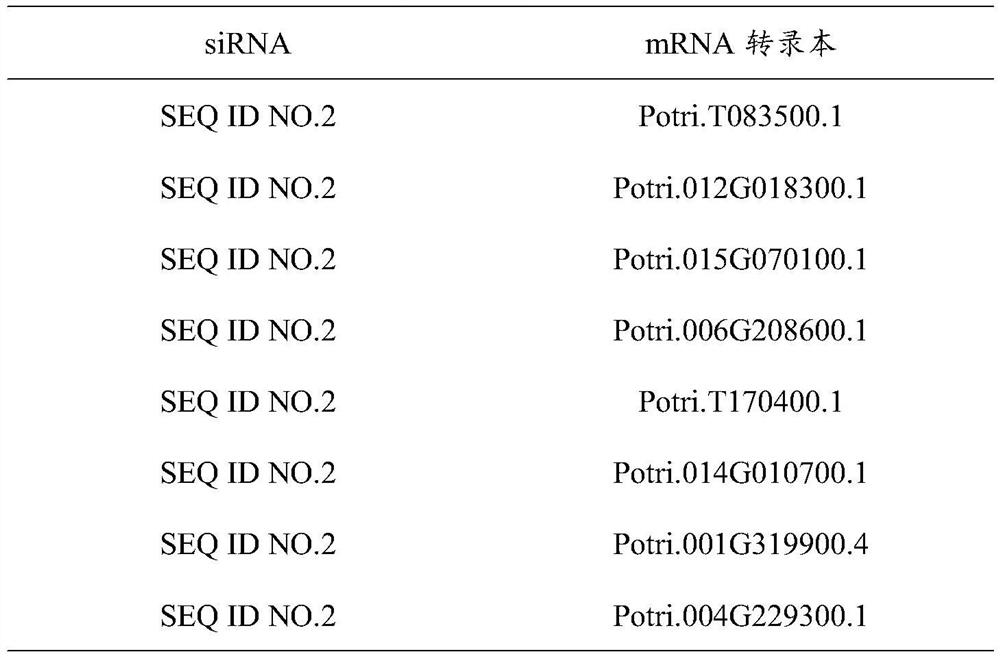
Method for identifying transcription regulation relationship of plant endogenous siRNA
An identification method and transcriptional regulation technology, applied in the field of molecular genetics, can solve problems such as low efficiency, no analysis method, and inability to perform high-throughput analysis, and achieve the effect of overcoming cumbersome, important theoretical significance and practical value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Experimental materials: Populus tomentosa annual clone '1316' was obtained from the National Populus tomentosa Germplasm Bank;
[0047] The specific operation steps are as follows:
[0048] Select individual leaves of Populus tomentosa for RNA extraction for small RNA sequencing, perform quality control on the original reads obtained by sequencing, and remove adapter sequences and low-quality sequences (including ambiguous base N, sequences with base quality less than 10 and length less than 18nt ) to get clean reads. CLC genomics_workbench 5.5 software was used to align the preprocessed sequences with the Sanger miRBase database (19.0): base mismatches were not allowed during the alignment process. In addition, it will be compared with the data in other non-coding databases ncRNA, piRNA, and Rfam: 2 base mismatches with the target sequence are allowed, and 2 bases are allowed to be shortened or extended from both ends of the target sequence. In the clean reads, the s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com